library(here) # manage file paths
library(socviz) # data and some useful functions
library(tidyverse) # your friend and mineTables with dplyr
Modern Plain Text Social Science: Week 5
October 7, 2024
dplyr is your toolkit for tabular data
So let’s
play with
some data
Load our libraries
Tidyverse components, again
library(tidyverse)Loading tidyverse: ggplot2Loading tidyverse: tibbleLoading tidyverse: tidyrLoading tidyverse: readrLoading tidyverse: purrrLoading tidyverse: dplyr
- Call the package and …
<|Draw graphs<|Nicer data tables<|Tidy your data<|Get data into R<|Fancy Iteration<|Action verbs for tables
Other tidyverse components
forcatshavenlubridatereadxlstringrreprex
<|Deal with factors<|Import Stata, SPSS, etc<|Dates, Durations, Times<|Import from spreadsheets<|Strings and Regular Expressions<|Make reproducible examples
Not all of these are attached when we do library(tidyverse)
dplyr lets you work with tibbles
- Remember, tibbles are tables of data where the columns can be of different types, such as numeric, logical, character, factor, etc.
- We’ll use dplyr to transform and summarize our data.
- The core logic is: split, apply, combine:
- Split or group the data into pieces
- Apply a function, i.e. act on the data in some way
- Combine the results back into a table
- We’ll use the pipe operator,
|>, to chain together sequences of actions on our tables.
dplyr’s core verbs
dplyr draws on the logic and language of database queries
Some actions to take on a single table
Subset either the rows or columns of or table—i.e. remove them before doing anything.
Group the data at the level we want, such as “By Race”, or “By Country”, or “By Religion within Regions” or “By Grade within Schools”.
Mutate the data. That is, change something at the current level of grouping. Mutating adds new columns to the table, or changes the content of an existing column. It never changes the number of rows.
Summarize or aggregate the data. That is, make something new at a higher level of grouping. E.g., calculate means or counts by some grouping variable. This will generally result in a smaller, summary table. Usually this will have the same number of rows as there are groups being summarized.
For each action there’s a function
- Subset has one function for rows and one for columns. We
filter()rows andselect()columns. - Group using
group_by(). - Mutate tables (i.e. add new columns, or re-make existing ones) using
mutate(). - Summarize tables (i.e. perform aggregating calculations) using
summarize().
Group and Summarize
General Social Survey data: gss_sm
# A tibble: 2,867 × 32
year id ballot age childs sibs degree race sex region income16
<dbl> <dbl> <labelled> <dbl> <dbl> <labe> <fct> <fct> <fct> <fct> <fct>
1 2016 1 1 47 3 2 Bache… White Male New E… $170000…
2 2016 2 2 61 0 3 High … White Male New E… $50000 …
3 2016 3 3 72 2 3 Bache… White Male New E… $75000 …
4 2016 4 1 43 4 3 High … White Fema… New E… $170000…
5 2016 5 3 55 2 2 Gradu… White Fema… New E… $170000…
6 2016 6 2 53 2 2 Junio… White Fema… New E… $60000 …
7 2016 7 1 50 2 2 High … White Male New E… $170000…
8 2016 8 3 23 3 6 High … Other Fema… Middl… $30000 …
9 2016 9 1 45 3 5 High … Black Male Middl… $60000 …
10 2016 10 3 71 4 1 Junio… White Male Middl… $60000 …
# ℹ 2,857 more rows
# ℹ 21 more variables: relig <fct>, marital <fct>, padeg <fct>, madeg <fct>,
# partyid <fct>, polviews <fct>, happy <fct>, partners <fct>, grass <fct>,
# zodiac <fct>, pres12 <labelled>, wtssall <dbl>, income_rc <fct>,
# agegrp <fct>, ageq <fct>, siblings <fct>, kids <fct>, religion <fct>,
# bigregion <fct>, partners_rc <fct>, obama <dbl>Notice how the tibble already tells us a lot.
Summarizing a Table
- Here’s what we’re going to do:
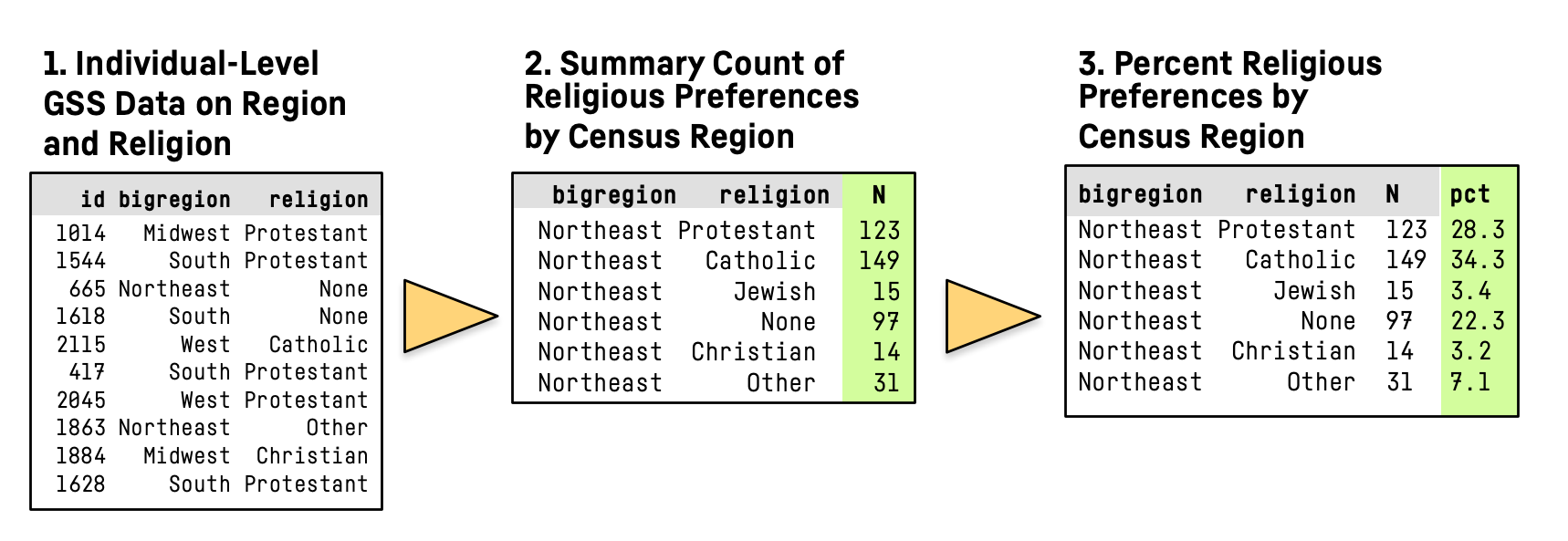 ]
]
Summarizing a Table
# A tibble: 2,867 × 3
id bigregion religion
<dbl> <fct> <fct>
1 1 Northeast None
2 2 Northeast None
3 3 Northeast Catholic
4 4 Northeast Catholic
5 5 Northeast None
6 6 Northeast None
7 7 Northeast None
8 8 Northeast Catholic
9 9 Northeast Protestant
10 10 Northeast None
# ℹ 2,857 more rowsWe’re just taking a look at the relevant columns here.
Group by one column or variable
# A tibble: 2,867 × 32
# Groups: bigregion [4]
year id ballot age childs sibs degree race sex region income16
<dbl> <dbl> <labelled> <dbl> <dbl> <labe> <fct> <fct> <fct> <fct> <fct>
1 2016 1 1 47 3 2 Bache… White Male New E… $170000…
2 2016 2 2 61 0 3 High … White Male New E… $50000 …
3 2016 3 3 72 2 3 Bache… White Male New E… $75000 …
4 2016 4 1 43 4 3 High … White Fema… New E… $170000…
5 2016 5 3 55 2 2 Gradu… White Fema… New E… $170000…
6 2016 6 2 53 2 2 Junio… White Fema… New E… $60000 …
7 2016 7 1 50 2 2 High … White Male New E… $170000…
8 2016 8 3 23 3 6 High … Other Fema… Middl… $30000 …
9 2016 9 1 45 3 5 High … Black Male Middl… $60000 …
10 2016 10 3 71 4 1 Junio… White Male Middl… $60000 …
# ℹ 2,857 more rows
# ℹ 21 more variables: relig <fct>, marital <fct>, padeg <fct>, madeg <fct>,
# partyid <fct>, polviews <fct>, happy <fct>, partners <fct>, grass <fct>,
# zodiac <fct>, pres12 <labelled>, wtssall <dbl>, income_rc <fct>,
# agegrp <fct>, ageq <fct>, siblings <fct>, kids <fct>, religion <fct>,
# bigregion <fct>, partners_rc <fct>, obama <dbl>Grouping just changes the logical structure of the tibble.
Group and summarize by one column
# A tibble: 2,867 × 32
year id ballot age childs sibs degree race sex region income16
<dbl> <dbl> <labelled> <dbl> <dbl> <labe> <fct> <fct> <fct> <fct> <fct>
1 2016 1 1 47 3 2 Bache… White Male New E… $170000…
2 2016 2 2 61 0 3 High … White Male New E… $50000 …
3 2016 3 3 72 2 3 Bache… White Male New E… $75000 …
4 2016 4 1 43 4 3 High … White Fema… New E… $170000…
5 2016 5 3 55 2 2 Gradu… White Fema… New E… $170000…
6 2016 6 2 53 2 2 Junio… White Fema… New E… $60000 …
7 2016 7 1 50 2 2 High … White Male New E… $170000…
8 2016 8 3 23 3 6 High … Other Fema… Middl… $30000 …
9 2016 9 1 45 3 5 High … Black Male Middl… $60000 …
10 2016 10 3 71 4 1 Junio… White Male Middl… $60000 …
# ℹ 2,857 more rows
# ℹ 21 more variables: relig <fct>, marital <fct>, padeg <fct>, madeg <fct>,
# partyid <fct>, polviews <fct>, happy <fct>, partners <fct>, grass <fct>,
# zodiac <fct>, pres12 <labelled>, wtssall <dbl>, income_rc <fct>,
# agegrp <fct>, ageq <fct>, siblings <fct>, kids <fct>, religion <fct>,
# bigregion <fct>, partners_rc <fct>, obama <dbl>Group and summarize by one column
# A tibble: 2,867 × 32
# Groups: bigregion [4]
year id ballot age childs sibs degree race sex region income16
<dbl> <dbl> <labelled> <dbl> <dbl> <labe> <fct> <fct> <fct> <fct> <fct>
1 2016 1 1 47 3 2 Bache… White Male New E… $170000…
2 2016 2 2 61 0 3 High … White Male New E… $50000 …
3 2016 3 3 72 2 3 Bache… White Male New E… $75000 …
4 2016 4 1 43 4 3 High … White Fema… New E… $170000…
5 2016 5 3 55 2 2 Gradu… White Fema… New E… $170000…
6 2016 6 2 53 2 2 Junio… White Fema… New E… $60000 …
7 2016 7 1 50 2 2 High … White Male New E… $170000…
8 2016 8 3 23 3 6 High … Other Fema… Middl… $30000 …
9 2016 9 1 45 3 5 High … Black Male Middl… $60000 …
10 2016 10 3 71 4 1 Junio… White Male Middl… $60000 …
# ℹ 2,857 more rows
# ℹ 21 more variables: relig <fct>, marital <fct>, padeg <fct>, madeg <fct>,
# partyid <fct>, polviews <fct>, happy <fct>, partners <fct>, grass <fct>,
# zodiac <fct>, pres12 <labelled>, wtssall <dbl>, income_rc <fct>,
# agegrp <fct>, ageq <fct>, siblings <fct>, kids <fct>, religion <fct>,
# bigregion <fct>, partners_rc <fct>, obama <dbl>Group and summarize by one column
# A tibble: 4 × 2
bigregion total
<fct> <int>
1 Northeast 488
2 Midwest 695
3 South 1052
4 West 632- The function
n()counts up the rows within each group.
- All the other columns are dropped in the summary operation
- Your original
gss_smtable is untouched
Group and summarize by two columns
# A tibble: 2,867 × 32
year id ballot age childs sibs degree race sex region income16
<dbl> <dbl> <labelled> <dbl> <dbl> <labe> <fct> <fct> <fct> <fct> <fct>
1 2016 1 1 47 3 2 Bache… White Male New E… $170000…
2 2016 2 2 61 0 3 High … White Male New E… $50000 …
3 2016 3 3 72 2 3 Bache… White Male New E… $75000 …
4 2016 4 1 43 4 3 High … White Fema… New E… $170000…
5 2016 5 3 55 2 2 Gradu… White Fema… New E… $170000…
6 2016 6 2 53 2 2 Junio… White Fema… New E… $60000 …
7 2016 7 1 50 2 2 High … White Male New E… $170000…
8 2016 8 3 23 3 6 High … Other Fema… Middl… $30000 …
9 2016 9 1 45 3 5 High … Black Male Middl… $60000 …
10 2016 10 3 71 4 1 Junio… White Male Middl… $60000 …
# ℹ 2,857 more rows
# ℹ 21 more variables: relig <fct>, marital <fct>, padeg <fct>, madeg <fct>,
# partyid <fct>, polviews <fct>, happy <fct>, partners <fct>, grass <fct>,
# zodiac <fct>, pres12 <labelled>, wtssall <dbl>, income_rc <fct>,
# agegrp <fct>, ageq <fct>, siblings <fct>, kids <fct>, religion <fct>,
# bigregion <fct>, partners_rc <fct>, obama <dbl>Group and summarize by two columns
# A tibble: 2,867 × 32
# Groups: bigregion, religion [24]
year id ballot age childs sibs degree race sex region income16
<dbl> <dbl> <labelled> <dbl> <dbl> <labe> <fct> <fct> <fct> <fct> <fct>
1 2016 1 1 47 3 2 Bache… White Male New E… $170000…
2 2016 2 2 61 0 3 High … White Male New E… $50000 …
3 2016 3 3 72 2 3 Bache… White Male New E… $75000 …
4 2016 4 1 43 4 3 High … White Fema… New E… $170000…
5 2016 5 3 55 2 2 Gradu… White Fema… New E… $170000…
6 2016 6 2 53 2 2 Junio… White Fema… New E… $60000 …
7 2016 7 1 50 2 2 High … White Male New E… $170000…
8 2016 8 3 23 3 6 High … Other Fema… Middl… $30000 …
9 2016 9 1 45 3 5 High … Black Male Middl… $60000 …
10 2016 10 3 71 4 1 Junio… White Male Middl… $60000 …
# ℹ 2,857 more rows
# ℹ 21 more variables: relig <fct>, marital <fct>, padeg <fct>, madeg <fct>,
# partyid <fct>, polviews <fct>, happy <fct>, partners <fct>, grass <fct>,
# zodiac <fct>, pres12 <labelled>, wtssall <dbl>, income_rc <fct>,
# agegrp <fct>, ageq <fct>, siblings <fct>, kids <fct>, religion <fct>,
# bigregion <fct>, partners_rc <fct>, obama <dbl>Group and summarize by two columns
# A tibble: 24 × 3
# Groups: bigregion [4]
bigregion religion total
<fct> <fct> <int>
1 Northeast Protestant 158
2 Northeast Catholic 162
3 Northeast Jewish 27
4 Northeast None 112
5 Northeast Other 28
6 Northeast <NA> 1
7 Midwest Protestant 325
8 Midwest Catholic 172
9 Midwest Jewish 3
10 Midwest None 157
# ℹ 14 more rows- The function
n()counts up the rows within the innermost (i.e. the rightmost) group.
Calculate frequencies
# A tibble: 2,867 × 32
year id ballot age childs sibs degree race sex region income16
<dbl> <dbl> <labelled> <dbl> <dbl> <labe> <fct> <fct> <fct> <fct> <fct>
1 2016 1 1 47 3 2 Bache… White Male New E… $170000…
2 2016 2 2 61 0 3 High … White Male New E… $50000 …
3 2016 3 3 72 2 3 Bache… White Male New E… $75000 …
4 2016 4 1 43 4 3 High … White Fema… New E… $170000…
5 2016 5 3 55 2 2 Gradu… White Fema… New E… $170000…
6 2016 6 2 53 2 2 Junio… White Fema… New E… $60000 …
7 2016 7 1 50 2 2 High … White Male New E… $170000…
8 2016 8 3 23 3 6 High … Other Fema… Middl… $30000 …
9 2016 9 1 45 3 5 High … Black Male Middl… $60000 …
10 2016 10 3 71 4 1 Junio… White Male Middl… $60000 …
# ℹ 2,857 more rows
# ℹ 21 more variables: relig <fct>, marital <fct>, padeg <fct>, madeg <fct>,
# partyid <fct>, polviews <fct>, happy <fct>, partners <fct>, grass <fct>,
# zodiac <fct>, pres12 <labelled>, wtssall <dbl>, income_rc <fct>,
# agegrp <fct>, ageq <fct>, siblings <fct>, kids <fct>, religion <fct>,
# bigregion <fct>, partners_rc <fct>, obama <dbl>Calculate frequencies
# A tibble: 2,867 × 32
# Groups: bigregion, religion [24]
year id ballot age childs sibs degree race sex region income16
<dbl> <dbl> <labelled> <dbl> <dbl> <labe> <fct> <fct> <fct> <fct> <fct>
1 2016 1 1 47 3 2 Bache… White Male New E… $170000…
2 2016 2 2 61 0 3 High … White Male New E… $50000 …
3 2016 3 3 72 2 3 Bache… White Male New E… $75000 …
4 2016 4 1 43 4 3 High … White Fema… New E… $170000…
5 2016 5 3 55 2 2 Gradu… White Fema… New E… $170000…
6 2016 6 2 53 2 2 Junio… White Fema… New E… $60000 …
7 2016 7 1 50 2 2 High … White Male New E… $170000…
8 2016 8 3 23 3 6 High … Other Fema… Middl… $30000 …
9 2016 9 1 45 3 5 High … Black Male Middl… $60000 …
10 2016 10 3 71 4 1 Junio… White Male Middl… $60000 …
# ℹ 2,857 more rows
# ℹ 21 more variables: relig <fct>, marital <fct>, padeg <fct>, madeg <fct>,
# partyid <fct>, polviews <fct>, happy <fct>, partners <fct>, grass <fct>,
# zodiac <fct>, pres12 <labelled>, wtssall <dbl>, income_rc <fct>,
# agegrp <fct>, ageq <fct>, siblings <fct>, kids <fct>, religion <fct>,
# bigregion <fct>, partners_rc <fct>, obama <dbl>Calculate frequencies
# A tibble: 24 × 3
# Groups: bigregion [4]
bigregion religion total
<fct> <fct> <int>
1 Northeast Protestant 158
2 Northeast Catholic 162
3 Northeast Jewish 27
4 Northeast None 112
5 Northeast Other 28
6 Northeast <NA> 1
7 Midwest Protestant 325
8 Midwest Catholic 172
9 Midwest Jewish 3
10 Midwest None 157
# ℹ 14 more rowsCalculate frequencies
# A tibble: 24 × 5
# Groups: bigregion [4]
bigregion religion total freq pct
<fct> <fct> <int> <dbl> <dbl>
1 Northeast Protestant 158 0.324 32.4
2 Northeast Catholic 162 0.332 33.2
3 Northeast Jewish 27 0.0553 5.5
4 Northeast None 112 0.230 23
5 Northeast Other 28 0.0574 5.7
6 Northeast <NA> 1 0.00205 0.2
7 Midwest Protestant 325 0.468 46.8
8 Midwest Catholic 172 0.247 24.7
9 Midwest Jewish 3 0.00432 0.4
10 Midwest None 157 0.226 22.6
# ℹ 14 more rows- The function
n()counts up the rows - Which rows? The ones fed down the pipeline
- The innermost (i.e. the rightmost) group.
Pipelines carry assumptions forward
gss_sm |>
group_by(bigregion, religion) |>
summarize(total = n()) |>
mutate(freq = total / sum(total),
pct = round((freq*100), 1))# A tibble: 24 × 5
# Groups: bigregion [4]
bigregion religion total freq pct
<fct> <fct> <int> <dbl> <dbl>
1 Northeast Protestant 158 0.324 32.4
2 Northeast Catholic 162 0.332 33.2
3 Northeast Jewish 27 0.0553 5.5
4 Northeast None 112 0.230 23
5 Northeast Other 28 0.0574 5.7
6 Northeast <NA> 1 0.00205 0.2
7 Midwest Protestant 325 0.468 46.8
8 Midwest Catholic 172 0.247 24.7
9 Midwest Jewish 3 0.00432 0.4
10 Midwest None 157 0.226 22.6
# ℹ 14 more rowsGroups are carried forward till summarized or explicitly ungrouped. Summary calculations are done on the innermost group, which “disappears”. (It becomes the rows in the summary table.)
Pipelines carry assumptions forward
gss_sm |>
group_by(bigregion, religion) |>
summarize(total = n()) |>
mutate(freq = total / sum(total),
pct = round((freq*100), 1)) # A tibble: 24 × 5
# Groups: bigregion [4]
bigregion religion total freq pct
<fct> <fct> <int> <dbl> <dbl>
1 Northeast Protestant 158 0.324 32.4
2 Northeast Catholic 162 0.332 33.2
3 Northeast Jewish 27 0.0553 5.5
4 Northeast None 112 0.230 23
5 Northeast Other 28 0.0574 5.7
6 Northeast <NA> 1 0.00205 0.2
7 Midwest Protestant 325 0.468 46.8
8 Midwest Catholic 172 0.247 24.7
9 Midwest Jewish 3 0.00432 0.4
10 Midwest None 157 0.226 22.6
# ℹ 14 more rowsmutate() is quite clever. See how we can immediately use freq, even though we are creating it in the same mutate() expression.
Convenience functions
gss_sm |>
group_by(bigregion, religion) |>
summarize(total = n()) |>
mutate(freq = total / sum(total),
pct = round((freq*100), 1)) # A tibble: 24 × 5
# Groups: bigregion [4]
bigregion religion total freq pct
<fct> <fct> <int> <dbl> <dbl>
1 Northeast Protestant 158 0.324 32.4
2 Northeast Catholic 162 0.332 33.2
3 Northeast Jewish 27 0.0553 5.5
4 Northeast None 112 0.230 23
5 Northeast Other 28 0.0574 5.7
6 Northeast <NA> 1 0.00205 0.2
7 Midwest Protestant 325 0.468 46.8
8 Midwest Catholic 172 0.247 24.7
9 Midwest Jewish 3 0.00432 0.4
10 Midwest None 157 0.226 22.6
# ℹ 14 more rowsWe’re going to be doing this group_by() … n() step a lot. Some shorthand for it would be useful.
Three options for counting up rows
- Use
n()
# A tibble: 24 × 3
# Groups: bigregion [4]
bigregion religion n
<fct> <fct> <int>
1 Northeast Protestant 158
2 Northeast Catholic 162
3 Northeast Jewish 27
4 Northeast None 112
5 Northeast Other 28
6 Northeast <NA> 1
7 Midwest Protestant 325
8 Midwest Catholic 172
9 Midwest Jewish 3
10 Midwest None 157
# ℹ 14 more rows- Group it yourself; result is grouped.
- Use
tally()
# A tibble: 24 × 3
# Groups: bigregion [4]
bigregion religion n
<fct> <fct> <int>
1 Northeast Protestant 158
2 Northeast Catholic 162
3 Northeast Jewish 27
4 Northeast None 112
5 Northeast Other 28
6 Northeast <NA> 1
7 Midwest Protestant 325
8 Midwest Catholic 172
9 Midwest Jewish 3
10 Midwest None 157
# ℹ 14 more rows- More compact; result is grouped.
- Use
count()
# A tibble: 24 × 3
bigregion religion n
<fct> <fct> <int>
1 Northeast Protestant 158
2 Northeast Catholic 162
3 Northeast Jewish 27
4 Northeast None 112
5 Northeast Other 28
6 Northeast <NA> 1
7 Midwest Protestant 325
8 Midwest Catholic 172
9 Midwest Jewish 3
10 Midwest None 157
# ℹ 14 more rows- One step; result is not grouped.
Pass results on to … a table
| religion | Northeast | Midwest | South | West |
|---|---|---|---|---|
| Protestant | 158 | 325 | 650 | 238 |
| Catholic | 162 | 172 | 160 | 155 |
| Jewish | 27 | 3 | 11 | 10 |
| None | 112 | 157 | 170 | 180 |
| Other | 28 | 33 | 50 | 48 |
| NA | 1 | 5 | 11 | 1 |
- More on
pivot_wider()soon …
Pass results on to … a graph

Pass results on to … an object
- You can do it like this …
rel_by_region <- gss_sm |>
count(bigregion, religion) |>
mutate(pct = round((n/sum(n))*100, 1))
rel_by_region# A tibble: 24 × 4
bigregion religion n pct
<fct> <fct> <int> <dbl>
1 Northeast Protestant 158 5.5
2 Northeast Catholic 162 5.7
3 Northeast Jewish 27 0.9
4 Northeast None 112 3.9
5 Northeast Other 28 1
6 Northeast <NA> 1 0
7 Midwest Protestant 325 11.3
8 Midwest Catholic 172 6
9 Midwest Jewish 3 0.1
10 Midwest None 157 5.5
# ℹ 14 more rowsPass results on to … an object
- You can do it like this …
rel_by_region <- gss_sm |>
count(bigregion, religion) |>
mutate(pct = round((n/sum(n))*100, 1))
rel_by_region# A tibble: 24 × 4
bigregion religion n pct
<fct> <fct> <int> <dbl>
1 Northeast Protestant 158 5.5
2 Northeast Catholic 162 5.7
3 Northeast Jewish 27 0.9
4 Northeast None 112 3.9
5 Northeast Other 28 1
6 Northeast <NA> 1 0
7 Midwest Protestant 325 11.3
8 Midwest Catholic 172 6
9 Midwest Jewish 3 0.1
10 Midwest None 157 5.5
# ℹ 14 more rows- Or like this!
gss_sm |>
count(bigregion, religion) |>
mutate(pct = round((n/sum(n))*100, 1)) ->
rel_by_region
rel_by_region# A tibble: 24 × 4
bigregion religion n pct
<fct> <fct> <int> <dbl>
1 Northeast Protestant 158 5.5
2 Northeast Catholic 162 5.7
3 Northeast Jewish 27 0.9
4 Northeast None 112 3.9
5 Northeast Other 28 1
6 Northeast <NA> 1 0
7 Midwest Protestant 325 11.3
8 Midwest Catholic 172 6
9 Midwest Jewish 3 0.1
10 Midwest None 157 5.5
# ℹ 14 more rowsRight assignmment is a thing, like Left
- Left assignment is standard
- This may feel awkward with a pipe: “
gss_tabgets the output of the following pipeline.”
Check by summarizing
rel_by_region <- gss_sm |>
count(bigregion, religion) |>
mutate(pct = round((n/sum(n))*100, 1))
rel_by_region# A tibble: 24 × 4
bigregion religion n pct
<fct> <fct> <int> <dbl>
1 Northeast Protestant 158 5.5
2 Northeast Catholic 162 5.7
3 Northeast Jewish 27 0.9
4 Northeast None 112 3.9
5 Northeast Other 28 1
6 Northeast <NA> 1 0
7 Midwest Protestant 325 11.3
8 Midwest Catholic 172 6
9 Midwest Jewish 3 0.1
10 Midwest None 157 5.5
# ℹ 14 more rowsHm, did I sum over right group?
Check by summarizing
rel_by_region <- gss_sm |>
count(bigregion, religion) |>
mutate(pct = round((n/sum(n))*100, 1))
rel_by_region# A tibble: 24 × 4
bigregion religion n pct
<fct> <fct> <int> <dbl>
1 Northeast Protestant 158 5.5
2 Northeast Catholic 162 5.7
3 Northeast Jewish 27 0.9
4 Northeast None 112 3.9
5 Northeast Other 28 1
6 Northeast <NA> 1 0
7 Midwest Protestant 325 11.3
8 Midwest Catholic 172 6
9 Midwest Jewish 3 0.1
10 Midwest None 157 5.5
# ℹ 14 more rowsHm, did I sum over right group?
Check by summarizing
count() returns ungrouped results, so there are no groups carry forward to the mutate() step.
With count(), the pct values here are the marginals for the whole table.
Check by summarizing
count() returns ungrouped results, so there are no groups carry forward to the mutate() step.
With count(), the pct values here are the marginals for the whole table.
# A tibble: 4 × 2
bigregion total
<fct> <dbl>
1 Northeast 100
2 Midwest 99.9
3 South 100
4 West 100. We get some rounding error because we used round() after summing originally.
Two lessons
Check your tables!
- Pipelines feed their content forward, so you need to make sure your results are not incorrect.
- Often, complex tables and graphs can be disturbingly plausible even when wrong.
- So, figure out what the result should be and test it!
- Starting with simple or toy cases can help with this process.
Two lessons
Inspect your pipes!
- Understand pipelines by running them forward or peeling them back a step at a time.
- This is a very effective way to understand your own and other people’s code.
Another example
Following a pipeline
gss_sm |>
group_by(race, sex, degree) |>
summarize(n = n(),
mean_age = mean(age, na.rm = TRUE),
mean_kids = mean(childs, na.rm = TRUE)) |>
mutate(pct = n/sum(n)*100) |>
filter(race !="Other") |>
drop_na() |>
ggplot(mapping = aes(x = mean_kids, y = degree)) + # Some ggplot ...
geom_col() + facet_grid(sex ~ race) +
labs(x = "Average number of Children", y = NULL)
Following a pipeline
# A tibble: 2,867 × 32
year id ballot age childs sibs degree race sex region income16
<dbl> <dbl> <labelled> <dbl> <dbl> <labe> <fct> <fct> <fct> <fct> <fct>
1 2016 1 1 47 3 2 Bache… White Male New E… $170000…
2 2016 2 2 61 0 3 High … White Male New E… $50000 …
3 2016 3 3 72 2 3 Bache… White Male New E… $75000 …
4 2016 4 1 43 4 3 High … White Fema… New E… $170000…
5 2016 5 3 55 2 2 Gradu… White Fema… New E… $170000…
6 2016 6 2 53 2 2 Junio… White Fema… New E… $60000 …
7 2016 7 1 50 2 2 High … White Male New E… $170000…
8 2016 8 3 23 3 6 High … Other Fema… Middl… $30000 …
9 2016 9 1 45 3 5 High … Black Male Middl… $60000 …
10 2016 10 3 71 4 1 Junio… White Male Middl… $60000 …
# ℹ 2,857 more rows
# ℹ 21 more variables: relig <fct>, marital <fct>, padeg <fct>, madeg <fct>,
# partyid <fct>, polviews <fct>, happy <fct>, partners <fct>, grass <fct>,
# zodiac <fct>, pres12 <labelled>, wtssall <dbl>, income_rc <fct>,
# agegrp <fct>, ageq <fct>, siblings <fct>, kids <fct>, religion <fct>,
# bigregion <fct>, partners_rc <fct>, obama <dbl>Following a pipeline
# A tibble: 2,867 × 32
# Groups: race, sex, degree [34]
year id ballot age childs sibs degree race sex region income16
<dbl> <dbl> <labelled> <dbl> <dbl> <labe> <fct> <fct> <fct> <fct> <fct>
1 2016 1 1 47 3 2 Bache… White Male New E… $170000…
2 2016 2 2 61 0 3 High … White Male New E… $50000 …
3 2016 3 3 72 2 3 Bache… White Male New E… $75000 …
4 2016 4 1 43 4 3 High … White Fema… New E… $170000…
5 2016 5 3 55 2 2 Gradu… White Fema… New E… $170000…
6 2016 6 2 53 2 2 Junio… White Fema… New E… $60000 …
7 2016 7 1 50 2 2 High … White Male New E… $170000…
8 2016 8 3 23 3 6 High … Other Fema… Middl… $30000 …
9 2016 9 1 45 3 5 High … Black Male Middl… $60000 …
10 2016 10 3 71 4 1 Junio… White Male Middl… $60000 …
# ℹ 2,857 more rows
# ℹ 21 more variables: relig <fct>, marital <fct>, padeg <fct>, madeg <fct>,
# partyid <fct>, polviews <fct>, happy <fct>, partners <fct>, grass <fct>,
# zodiac <fct>, pres12 <labelled>, wtssall <dbl>, income_rc <fct>,
# agegrp <fct>, ageq <fct>, siblings <fct>, kids <fct>, religion <fct>,
# bigregion <fct>, partners_rc <fct>, obama <dbl>Following a pipeline
# A tibble: 34 × 6
# Groups: race, sex [6]
race sex degree n mean_age mean_kids
<fct> <fct> <fct> <int> <dbl> <dbl>
1 White Male Lt High School 96 52.9 2.45
2 White Male High School 470 48.8 1.61
3 White Male Junior College 65 47.1 1.54
4 White Male Bachelor 208 48.6 1.35
5 White Male Graduate 112 56.0 1.71
6 White Female Lt High School 101 55.4 2.81
7 White Female High School 587 51.9 1.98
8 White Female Junior College 101 48.2 1.91
9 White Female Bachelor 218 49.2 1.44
10 White Female Graduate 138 53.6 1.38
# ℹ 24 more rowsFollowing a pipeline
# A tibble: 34 × 7
# Groups: race, sex [6]
race sex degree n mean_age mean_kids pct
<fct> <fct> <fct> <int> <dbl> <dbl> <dbl>
1 White Male Lt High School 96 52.9 2.45 10.1
2 White Male High School 470 48.8 1.61 49.4
3 White Male Junior College 65 47.1 1.54 6.83
4 White Male Bachelor 208 48.6 1.35 21.9
5 White Male Graduate 112 56.0 1.71 11.8
6 White Female Lt High School 101 55.4 2.81 8.79
7 White Female High School 587 51.9 1.98 51.1
8 White Female Junior College 101 48.2 1.91 8.79
9 White Female Bachelor 218 49.2 1.44 19.0
10 White Female Graduate 138 53.6 1.38 12.0
# ℹ 24 more rowsFollowing a pipeline
# A tibble: 23 × 7
# Groups: race, sex [4]
race sex degree n mean_age mean_kids pct
<fct> <fct> <fct> <int> <dbl> <dbl> <dbl>
1 White Male Lt High School 96 52.9 2.45 10.1
2 White Male High School 470 48.8 1.61 49.4
3 White Male Junior College 65 47.1 1.54 6.83
4 White Male Bachelor 208 48.6 1.35 21.9
5 White Male Graduate 112 56.0 1.71 11.8
6 White Female Lt High School 101 55.4 2.81 8.79
7 White Female High School 587 51.9 1.98 51.1
8 White Female Junior College 101 48.2 1.91 8.79
9 White Female Bachelor 218 49.2 1.44 19.0
10 White Female Graduate 138 53.6 1.38 12.0
# ℹ 13 more rowsFollowing a pipeline
# A tibble: 20 × 7
# Groups: race, sex [4]
race sex degree n mean_age mean_kids pct
<fct> <fct> <fct> <int> <dbl> <dbl> <dbl>
1 White Male Lt High School 96 52.9 2.45 10.1
2 White Male High School 470 48.8 1.61 49.4
3 White Male Junior College 65 47.1 1.54 6.83
4 White Male Bachelor 208 48.6 1.35 21.9
5 White Male Graduate 112 56.0 1.71 11.8
6 White Female Lt High School 101 55.4 2.81 8.79
7 White Female High School 587 51.9 1.98 51.1
8 White Female Junior College 101 48.2 1.91 8.79
9 White Female Bachelor 218 49.2 1.44 19.0
10 White Female Graduate 138 53.6 1.38 12.0
11 Black Male Lt High School 17 56.1 3 8.21
12 Black Male High School 142 43.6 1.96 68.6
13 Black Male Junior College 16 47.1 1.31 7.73
14 Black Male Bachelor 22 41.6 1.14 10.6
15 Black Male Graduate 8 53.1 1.88 3.86
16 Black Female Lt High School 43 51.0 2.91 15.2
17 Black Female High School 150 43.1 2.14 53.0
18 Black Female Junior College 17 45.8 1.82 6.01
19 Black Female Bachelor 49 47.0 1.76 17.3
20 Black Female Graduate 23 51.2 1.74 8.13Following a pipeline
# A tibble: 4 × 3
# Groups: race [2]
race sex grp_totpct
<fct> <fct> <dbl>
1 White Male 100
2 White Female 99.7
3 Black Male 99.0
4 Black Female 99.6Conditional selection
Conditionals in select() & filter()
- Some new data, this time on national rates of cadaveric organ donation:
# A tibble: 238 × 21
country year donors pop pop_dens gdp gdp_lag health health_lag
<chr> <date> <dbl> <int> <dbl> <int> <int> <dbl> <dbl>
1 Australia NA NA 17065 0.220 16774 16591 1300 1224
2 Australia 1991-01-01 12.1 17284 0.223 17171 16774 1379 1300
3 Australia 1992-01-01 12.4 17495 0.226 17914 17171 1455 1379
4 Australia 1993-01-01 12.5 17667 0.228 18883 17914 1540 1455
5 Australia 1994-01-01 10.2 17855 0.231 19849 18883 1626 1540
6 Australia 1995-01-01 10.2 18072 0.233 21079 19849 1737 1626
7 Australia 1996-01-01 10.6 18311 0.237 21923 21079 1846 1737
8 Australia 1997-01-01 10.3 18518 0.239 22961 21923 1948 1846
9 Australia 1998-01-01 10.5 18711 0.242 24148 22961 2077 1948
10 Australia 1999-01-01 8.67 18926 0.244 25445 24148 2231 2077
# ℹ 228 more rows
# ℹ 12 more variables: pubhealth <dbl>, roads <dbl>, cerebvas <int>,
# assault <int>, external <int>, txp_pop <dbl>, world <chr>, opt <chr>,
# consent_law <chr>, consent_practice <chr>, consistent <chr>, ccode <chr>Conditionals in select() & filter()
# A tibble: 30 × 21
country year donors pop pop_dens gdp gdp_lag health health_lag
<chr> <date> <dbl> <int> <dbl> <int> <int> <dbl> <dbl>
1 Canada 2000-01-01 15.3 30770 0.309 28472 26658 2541 2400
2 Denmark 1992-01-01 16.1 5171 12.0 19644 19126 1660 1603
3 Ireland 1991-01-01 19 3534 5.03 13495 12917 884 791
4 Ireland 1992-01-01 19.5 3558 5.06 14241 13495 1005 884
5 Ireland 1993-01-01 17.1 3576 5.09 14927 14241 1041 1005
6 Ireland 1994-01-01 20.3 3590 5.11 15990 14927 1119 1041
7 Ireland 1995-01-01 24.6 3609 5.14 17789 15990 1208 1119
8 Ireland 1996-01-01 16.8 3636 5.17 19245 17789 1269 1208
9 Ireland 1997-01-01 20.9 3673 5.23 22017 19245 1417 1269
10 Ireland 1998-01-01 23.8 3715 5.29 23995 22017 1487 1417
# ℹ 20 more rows
# ℹ 12 more variables: pubhealth <dbl>, roads <dbl>, cerebvas <int>,
# assault <int>, external <int>, txp_pop <dbl>, world <chr>, opt <chr>,
# consent_law <chr>, consent_practice <chr>, consistent <chr>, ccode <chr>Conditionals in select() & filter()
# A tibble: 238 × 8
country year pop gdp gdp_lag cerebvas assault external
<chr> <date> <int> <int> <int> <int> <int> <int>
1 Australia NA 17065 16774 16591 682 21 444
2 Australia 1991-01-01 17284 17171 16774 647 19 425
3 Australia 1992-01-01 17495 17914 17171 630 17 406
4 Australia 1993-01-01 17667 18883 17914 611 18 376
5 Australia 1994-01-01 17855 19849 18883 631 17 387
6 Australia 1995-01-01 18072 21079 19849 592 16 371
7 Australia 1996-01-01 18311 21923 21079 576 17 395
8 Australia 1997-01-01 18518 22961 21923 525 17 385
9 Australia 1998-01-01 18711 24148 22961 516 16 410
10 Australia 1999-01-01 18926 25445 24148 493 15 409
# ℹ 228 more rowsUse where() to test columns.
Conditionals in select() & filter()
When telling where() to use is.integer() to test each column, we don’t put parentheses at the end of its name. If we did, R would try to evaluate is.integer() right then, and fail:
> organdata |>
+ select(country, year, where(is.integer()))
Error: 0 arguments passed to 'is.integer' which requires 1
Run `rlang::last_error()` to see where the error occurred.This is true in similar situations elsewhere as well.
Conditionals in select() & filter()
# A tibble: 238 × 8
country year world opt consent_law consent_practice consistent ccode
<chr> <date> <chr> <chr> <chr> <chr> <chr> <chr>
1 Austral… NA Libe… In Informed Informed Yes Oz
2 Austral… 1991-01-01 Libe… In Informed Informed Yes Oz
3 Austral… 1992-01-01 Libe… In Informed Informed Yes Oz
4 Austral… 1993-01-01 Libe… In Informed Informed Yes Oz
5 Austral… 1994-01-01 Libe… In Informed Informed Yes Oz
6 Austral… 1995-01-01 Libe… In Informed Informed Yes Oz
7 Austral… 1996-01-01 Libe… In Informed Informed Yes Oz
8 Austral… 1997-01-01 Libe… In Informed Informed Yes Oz
9 Austral… 1998-01-01 Libe… In Informed Informed Yes Oz
10 Austral… 1999-01-01 Libe… In Informed Informed Yes Oz
# ℹ 228 more rowsWe have functions like e.g. is.character(), is.numeric(), is.logical(), is.factor(), etc. All return either TRUE or FALSE.
Conditionals in select() & filter()
Sometimes we don’t pass a function, but do want to use the result of one:
# A tibble: 238 × 4
country year gdp gdp_lag
<chr> <date> <int> <int>
1 Australia NA 16774 16591
2 Australia 1991-01-01 17171 16774
3 Australia 1992-01-01 17914 17171
4 Australia 1993-01-01 18883 17914
5 Australia 1994-01-01 19849 18883
6 Australia 1995-01-01 21079 19849
7 Australia 1996-01-01 21923 21079
8 Australia 1997-01-01 22961 21923
9 Australia 1998-01-01 24148 22961
10 Australia 1999-01-01 25445 24148
# ℹ 228 more rowsWe have starts_with(), ends_with(), contains(), matches(), and num_range(). Collectively these are “tidy selectors”.
Conditionals in select() & filter()
# A tibble: 28 × 21
country year donors pop pop_dens gdp gdp_lag health health_lag
<chr> <date> <dbl> <int> <dbl> <int> <int> <dbl> <dbl>
1 Australia NA NA 17065 0.220 16774 16591 1300 1224
2 Australia 1991-01-01 12.1 17284 0.223 17171 16774 1379 1300
3 Australia 1992-01-01 12.4 17495 0.226 17914 17171 1455 1379
4 Australia 1993-01-01 12.5 17667 0.228 18883 17914 1540 1455
5 Australia 1994-01-01 10.2 17855 0.231 19849 18883 1626 1540
6 Australia 1995-01-01 10.2 18072 0.233 21079 19849 1737 1626
7 Australia 1996-01-01 10.6 18311 0.237 21923 21079 1846 1737
8 Australia 1997-01-01 10.3 18518 0.239 22961 21923 1948 1846
9 Australia 1998-01-01 10.5 18711 0.242 24148 22961 2077 1948
10 Australia 1999-01-01 8.67 18926 0.244 25445 24148 2231 2077
# ℹ 18 more rows
# ℹ 12 more variables: pubhealth <dbl>, roads <dbl>, cerebvas <int>,
# assault <int>, external <int>, txp_pop <dbl>, world <chr>, opt <chr>,
# consent_law <chr>, consent_practice <chr>, consistent <chr>, ccode <chr>This could get cumbersome fast.
Use %in% for multiple selections
my_countries <- c("Australia", "Canada", "United States", "Ireland")
organdata |>
filter(country %in% my_countries) # A tibble: 56 × 21
country year donors pop pop_dens gdp gdp_lag health health_lag
<chr> <date> <dbl> <int> <dbl> <int> <int> <dbl> <dbl>
1 Australia NA NA 17065 0.220 16774 16591 1300 1224
2 Australia 1991-01-01 12.1 17284 0.223 17171 16774 1379 1300
3 Australia 1992-01-01 12.4 17495 0.226 17914 17171 1455 1379
4 Australia 1993-01-01 12.5 17667 0.228 18883 17914 1540 1455
5 Australia 1994-01-01 10.2 17855 0.231 19849 18883 1626 1540
6 Australia 1995-01-01 10.2 18072 0.233 21079 19849 1737 1626
7 Australia 1996-01-01 10.6 18311 0.237 21923 21079 1846 1737
8 Australia 1997-01-01 10.3 18518 0.239 22961 21923 1948 1846
9 Australia 1998-01-01 10.5 18711 0.242 24148 22961 2077 1948
10 Australia 1999-01-01 8.67 18926 0.244 25445 24148 2231 2077
# ℹ 46 more rows
# ℹ 12 more variables: pubhealth <dbl>, roads <dbl>, cerebvas <int>,
# assault <int>, external <int>, txp_pop <dbl>, world <chr>, opt <chr>,
# consent_law <chr>, consent_practice <chr>, consistent <chr>, ccode <chr>Negating %in%
my_countries <- c("Australia", "Canada", "United States", "Ireland")
organdata |>
filter(!(country %in% my_countries)) # A tibble: 182 × 21
country year donors pop pop_dens gdp gdp_lag health health_lag
<chr> <date> <dbl> <int> <dbl> <int> <int> <dbl> <dbl>
1 Austria NA NA 7678 9.16 18914 17425 1344 1255
2 Austria 1991-01-01 27.6 7755 9.25 19860 18914 1419 1344
3 Austria 1992-01-01 23.1 7841 9.35 20601 19860 1551 1419
4 Austria 1993-01-01 26.2 7906 9.43 21119 20601 1674 1551
5 Austria 1994-01-01 21.4 7936 9.46 21940 21119 1739 1674
6 Austria 1995-01-01 21.5 7948 9.48 22817 21940 1865 1739
7 Austria 1996-01-01 24.7 7959 9.49 23798 22817 1986 1865
8 Austria 1997-01-01 19.5 7968 9.50 24364 23798 1848 1986
9 Austria 1998-01-01 20.7 7977 9.51 25423 24364 1953 1848
10 Austria 1999-01-01 25.9 7992 9.53 26513 25423 2069 1953
# ℹ 172 more rows
# ℹ 12 more variables: pubhealth <dbl>, roads <dbl>, cerebvas <int>,
# assault <int>, external <int>, txp_pop <dbl>, world <chr>, opt <chr>,
# consent_law <chr>, consent_practice <chr>, consistent <chr>, ccode <chr>Also a bit awkward. There’s no built-in “Not in” operator.
A custom operator
# A tibble: 182 × 21
country year donors pop pop_dens gdp gdp_lag health health_lag
<chr> <date> <dbl> <int> <dbl> <int> <int> <dbl> <dbl>
1 Austria NA NA 7678 9.16 18914 17425 1344 1255
2 Austria 1991-01-01 27.6 7755 9.25 19860 18914 1419 1344
3 Austria 1992-01-01 23.1 7841 9.35 20601 19860 1551 1419
4 Austria 1993-01-01 26.2 7906 9.43 21119 20601 1674 1551
5 Austria 1994-01-01 21.4 7936 9.46 21940 21119 1739 1674
6 Austria 1995-01-01 21.5 7948 9.48 22817 21940 1865 1739
7 Austria 1996-01-01 24.7 7959 9.49 23798 22817 1986 1865
8 Austria 1997-01-01 19.5 7968 9.50 24364 23798 1848 1986
9 Austria 1998-01-01 20.7 7977 9.51 25423 24364 1953 1848
10 Austria 1999-01-01 25.9 7992 9.53 26513 25423 2069 1953
# ℹ 172 more rows
# ℹ 12 more variables: pubhealth <dbl>, roads <dbl>, cerebvas <int>,
# assault <int>, external <int>, txp_pop <dbl>, world <chr>, opt <chr>,
# consent_law <chr>, consent_practice <chr>, consistent <chr>, ccode <chr>Using across()
Do more than one thing
Earlier we saw this:
gss_sm |>
group_by(race, sex, degree) |>
summarize(n = n(),
mean_age = mean(age, na.rm = TRUE),
mean_kids = mean(childs, na.rm = TRUE))# A tibble: 34 × 6
# Groups: race, sex [6]
race sex degree n mean_age mean_kids
<fct> <fct> <fct> <int> <dbl> <dbl>
1 White Male Lt High School 96 52.9 2.45
2 White Male High School 470 48.8 1.61
3 White Male Junior College 65 47.1 1.54
4 White Male Bachelor 208 48.6 1.35
5 White Male Graduate 112 56.0 1.71
6 White Female Lt High School 101 55.4 2.81
7 White Female High School 587 51.9 1.98
8 White Female Junior College 101 48.2 1.91
9 White Female Bachelor 218 49.2 1.44
10 White Female Graduate 138 53.6 1.38
# ℹ 24 more rowsDo more than one thing
Similarly for organdata we might want to do:
organdata |>
group_by(consent_law, country) |>
summarize(donors_mean = mean(donors, na.rm = TRUE),
donors_sd = sd(donors, na.rm = TRUE),
gdp_mean = mean(gdp, na.rm = TRUE),
health_mean = mean(health, na.rm = TRUE),
roads_mean = mean(roads, na.rm = TRUE))# A tibble: 17 × 7
# Groups: consent_law [2]
consent_law country donors_mean donors_sd gdp_mean health_mean roads_mean
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Informed Australia 10.6 1.14 22179. 1958. 105.
2 Informed Canada 14.0 0.751 23711. 2272. 109.
3 Informed Denmark 13.1 1.47 23722. 2054. 102.
4 Informed Germany 13.0 0.611 22163. 2349. 113.
5 Informed Ireland 19.8 2.48 20824. 1480. 118.
6 Informed Netherlands 13.7 1.55 23013. 1993. 76.1
7 Informed United Kin… 13.5 0.775 21359. 1561. 67.9
8 Informed United Sta… 20.0 1.33 29212. 3988. 155.
9 Presumed Austria 23.5 2.42 23876. 1875. 150.
10 Presumed Belgium 21.9 1.94 22500. 1958. 155.
11 Presumed Finland 18.4 1.53 21019. 1615. 93.6
12 Presumed France 16.8 1.60 22603. 2160. 156.
13 Presumed Italy 11.1 4.28 21554. 1757 122.
14 Presumed Norway 15.4 1.11 26448. 2217. 70.0
15 Presumed Spain 28.1 4.96 16933 1289. 161.
16 Presumed Sweden 13.1 1.75 22415. 1951. 72.3
17 Presumed Switzerland 14.2 1.71 27233 2776. 96.4This works, but it’s really tedious. Also error-prone.
Use across()
Instead, use across() to apply a function to more than one column.
my_vars <- c("gdp", "donors", "roads")
## nested parens again, but it's worth it
organdata |>
group_by(consent_law, country) |>
summarize(across(all_of(my_vars),
list(avg = \(x) mean(x, na.rm = TRUE))
)
) # A tibble: 17 × 5
# Groups: consent_law [2]
consent_law country gdp_avg donors_avg roads_avg
<chr> <chr> <dbl> <dbl> <dbl>
1 Informed Australia 22179. 10.6 105.
2 Informed Canada 23711. 14.0 109.
3 Informed Denmark 23722. 13.1 102.
4 Informed Germany 22163. 13.0 113.
5 Informed Ireland 20824. 19.8 118.
6 Informed Netherlands 23013. 13.7 76.1
7 Informed United Kingdom 21359. 13.5 67.9
8 Informed United States 29212. 20.0 155.
9 Presumed Austria 23876. 23.5 150.
10 Presumed Belgium 22500. 21.9 155.
11 Presumed Finland 21019. 18.4 93.6
12 Presumed France 22603. 16.8 156.
13 Presumed Italy 21554. 11.1 122.
14 Presumed Norway 26448. 15.4 70.0
15 Presumed Spain 16933 28.1 161.
16 Presumed Sweden 22415. 13.1 72.3
17 Presumed Switzerland 27233 14.2 96.4Let’s look at that again
Let’s look at that again
# A tibble: 238 × 21
country year donors pop pop_dens gdp gdp_lag health health_lag
<chr> <date> <dbl> <int> <dbl> <int> <int> <dbl> <dbl>
1 Australia NA NA 17065 0.220 16774 16591 1300 1224
2 Australia 1991-01-01 12.1 17284 0.223 17171 16774 1379 1300
3 Australia 1992-01-01 12.4 17495 0.226 17914 17171 1455 1379
4 Australia 1993-01-01 12.5 17667 0.228 18883 17914 1540 1455
5 Australia 1994-01-01 10.2 17855 0.231 19849 18883 1626 1540
6 Australia 1995-01-01 10.2 18072 0.233 21079 19849 1737 1626
7 Australia 1996-01-01 10.6 18311 0.237 21923 21079 1846 1737
8 Australia 1997-01-01 10.3 18518 0.239 22961 21923 1948 1846
9 Australia 1998-01-01 10.5 18711 0.242 24148 22961 2077 1948
10 Australia 1999-01-01 8.67 18926 0.244 25445 24148 2231 2077
# ℹ 228 more rows
# ℹ 12 more variables: pubhealth <dbl>, roads <dbl>, cerebvas <int>,
# assault <int>, external <int>, txp_pop <dbl>, world <chr>, opt <chr>,
# consent_law <chr>, consent_practice <chr>, consistent <chr>, ccode <chr>Let’s look at that again
# A tibble: 238 × 21
# Groups: consent_law, country [17]
country year donors pop pop_dens gdp gdp_lag health health_lag
<chr> <date> <dbl> <int> <dbl> <int> <int> <dbl> <dbl>
1 Australia NA NA 17065 0.220 16774 16591 1300 1224
2 Australia 1991-01-01 12.1 17284 0.223 17171 16774 1379 1300
3 Australia 1992-01-01 12.4 17495 0.226 17914 17171 1455 1379
4 Australia 1993-01-01 12.5 17667 0.228 18883 17914 1540 1455
5 Australia 1994-01-01 10.2 17855 0.231 19849 18883 1626 1540
6 Australia 1995-01-01 10.2 18072 0.233 21079 19849 1737 1626
7 Australia 1996-01-01 10.6 18311 0.237 21923 21079 1846 1737
8 Australia 1997-01-01 10.3 18518 0.239 22961 21923 1948 1846
9 Australia 1998-01-01 10.5 18711 0.242 24148 22961 2077 1948
10 Australia 1999-01-01 8.67 18926 0.244 25445 24148 2231 2077
# ℹ 228 more rows
# ℹ 12 more variables: pubhealth <dbl>, roads <dbl>, cerebvas <int>,
# assault <int>, external <int>, txp_pop <dbl>, world <chr>, opt <chr>,
# consent_law <chr>, consent_practice <chr>, consistent <chr>, ccode <chr>Let’s look at that again
# A tibble: 17 × 5
# Groups: consent_law [2]
consent_law country gdp_avg donors_avg roads_avg
<chr> <chr> <dbl> <dbl> <dbl>
1 Informed Australia 22179. 10.6 105.
2 Informed Canada 23711. 14.0 109.
3 Informed Denmark 23722. 13.1 102.
4 Informed Germany 22163. 13.0 113.
5 Informed Ireland 20824. 19.8 118.
6 Informed Netherlands 23013. 13.7 76.1
7 Informed United Kingdom 21359. 13.5 67.9
8 Informed United States 29212. 20.0 155.
9 Presumed Austria 23876. 23.5 150.
10 Presumed Belgium 22500. 21.9 155.
11 Presumed Finland 21019. 18.4 93.6
12 Presumed France 22603. 16.8 156.
13 Presumed Italy 21554. 11.1 122.
14 Presumed Norway 26448. 15.4 70.0
15 Presumed Spain 16933 28.1 161.
16 Presumed Sweden 22415. 13.1 72.3
17 Presumed Switzerland 27233 14.2 96.4Let’s look at that again
my_varsare selected byacross()- We use
all_of()orany_of()to be explicit list()of the formresult = functiongives the new columns that will be calculated.- The thing inside the list with the “waving person”,
\(x), is an anonymous function
We can calculate more than one thing
my_vars <- c("gdp", "donors", "roads")
organdata |>
group_by(consent_law, country) |>
summarize(across(all_of(my_vars),
list(avg = \(x) mean(x, na.rm = TRUE),
sdev = \(x) sd(x, na.rm = TRUE),
md = \(x) median(x, na.rm = TRUE))
)
)# A tibble: 17 × 11
# Groups: consent_law [2]
consent_law country gdp_avg gdp_sdev gdp_md donors_avg donors_sdev donors_md
<chr> <chr> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
1 Informed Austral… 22179. 3959. 21923 10.6 1.14 10.4
2 Informed Canada 23711. 3966. 22764 14.0 0.751 14.0
3 Informed Denmark 23722. 3896. 23548 13.1 1.47 12.9
4 Informed Germany 22163. 2501. 22164 13.0 0.611 13
5 Informed Ireland 20824. 6670. 19245 19.8 2.48 19.2
6 Informed Netherl… 23013. 3770. 22541 13.7 1.55 13.8
7 Informed United … 21359. 3929. 20839 13.5 0.775 13.5
8 Informed United … 29212. 4571. 28772 20.0 1.33 20.1
9 Presumed Austria 23876. 3343. 23798 23.5 2.42 23.8
10 Presumed Belgium 22500. 3171. 22152 21.9 1.94 21.4
11 Presumed Finland 21019. 3668. 19842 18.4 1.53 19.4
12 Presumed France 22603. 3260. 21990 16.8 1.60 16.6
13 Presumed Italy 21554. 2781. 21396 11.1 4.28 11.3
14 Presumed Norway 26448. 6492. 26218 15.4 1.11 15.4
15 Presumed Spain 16933 2888. 16416 28.1 4.96 28
16 Presumed Sweden 22415. 3213. 22029 13.1 1.75 12.7
17 Presumed Switzer… 27233 2153. 26304 14.2 1.71 14.4
# ℹ 3 more variables: roads_avg <dbl>, roads_sdev <dbl>, roads_md <dbl>It’s OK to use the function names
my_vars <- c("gdp", "donors", "roads")
organdata |>
group_by(consent_law, country) |>
summarize(across(all_of(my_vars),
list(mean = \(x) mean(x, na.rm = TRUE),
sd = \(x) sd(x, na.rm = TRUE),
median = \(x) median(x, na.rm = TRUE))
)
)# A tibble: 17 × 11
# Groups: consent_law [2]
consent_law country gdp_mean gdp_sd gdp_median donors_mean donors_sd
<chr> <chr> <dbl> <dbl> <int> <dbl> <dbl>
1 Informed Australia 22179. 3959. 21923 10.6 1.14
2 Informed Canada 23711. 3966. 22764 14.0 0.751
3 Informed Denmark 23722. 3896. 23548 13.1 1.47
4 Informed Germany 22163. 2501. 22164 13.0 0.611
5 Informed Ireland 20824. 6670. 19245 19.8 2.48
6 Informed Netherlands 23013. 3770. 22541 13.7 1.55
7 Informed United Kingdom 21359. 3929. 20839 13.5 0.775
8 Informed United States 29212. 4571. 28772 20.0 1.33
9 Presumed Austria 23876. 3343. 23798 23.5 2.42
10 Presumed Belgium 22500. 3171. 22152 21.9 1.94
11 Presumed Finland 21019. 3668. 19842 18.4 1.53
12 Presumed France 22603. 3260. 21990 16.8 1.60
13 Presumed Italy 21554. 2781. 21396 11.1 4.28
14 Presumed Norway 26448. 6492. 26218 15.4 1.11
15 Presumed Spain 16933 2888. 16416 28.1 4.96
16 Presumed Sweden 22415. 3213. 22029 13.1 1.75
17 Presumed Switzerland 27233 2153. 26304 14.2 1.71
# ℹ 4 more variables: donors_median <dbl>, roads_mean <dbl>, roads_sd <dbl>,
# roads_median <dbl>Selection with across(where())
organdata |>
group_by(consent_law, country) |>
summarize(across(where(is.numeric),
list(mean = \(x) mean(x, na.rm = TRUE),
sd = \(x) sd(x, na.rm = TRUE),
median = \(x) median(x, na.rm = TRUE))
)
) |>
print(n = 3) # just to save slide space# A tibble: 17 × 41
# Groups: consent_law [2]
consent_law country donors_mean donors_sd donors_median pop_mean pop_sd
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Informed Australia 10.6 1.14 10.4 18318. 831.
2 Informed Canada 14.0 0.751 14.0 29608. 1193.
3 Informed Denmark 13.1 1.47 12.9 5257. 80.6
# ℹ 14 more rows
# ℹ 34 more variables: pop_median <int>, pop_dens_mean <dbl>,
# pop_dens_sd <dbl>, pop_dens_median <dbl>, gdp_mean <dbl>, gdp_sd <dbl>,
# gdp_median <int>, gdp_lag_mean <dbl>, gdp_lag_sd <dbl>,
# gdp_lag_median <dbl>, health_mean <dbl>, health_sd <dbl>,
# health_median <dbl>, health_lag_mean <dbl>, health_lag_sd <dbl>,
# health_lag_median <dbl>, pubhealth_mean <dbl>, pubhealth_sd <dbl>, …Name new columns with .names
organdata |>
group_by(consent_law, country) |>
summarize(across(where(is.numeric),
list(mean = \(x) mean(x, na.rm = TRUE),
sd = \(x) sd(x, na.rm = TRUE),
median = \(x) median(x, na.rm = TRUE))
),
.names = "{fn}_{col}") |>
print(n = 3) # A tibble: 17 × 42
# Groups: consent_law [2]
consent_law country donors_mean donors_sd donors_median pop_mean pop_sd
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Informed Australia 10.6 1.14 10.4 18318. 831.
2 Informed Canada 14.0 0.751 14.0 29608. 1193.
3 Informed Denmark 13.1 1.47 12.9 5257. 80.6
# ℹ 14 more rows
# ℹ 35 more variables: pop_median <int>, pop_dens_mean <dbl>,
# pop_dens_sd <dbl>, pop_dens_median <dbl>, gdp_mean <dbl>, gdp_sd <dbl>,
# gdp_median <int>, gdp_lag_mean <dbl>, gdp_lag_sd <dbl>,
# gdp_lag_median <dbl>, health_mean <dbl>, health_sd <dbl>,
# health_median <dbl>, health_lag_mean <dbl>, health_lag_sd <dbl>,
# health_lag_median <dbl>, pubhealth_mean <dbl>, pubhealth_sd <dbl>, …Name new columns with .names
In tidyverse functions, arguments that begin with a “.” generally have it in order to avoid confusion with existing items, or are “pronouns” referring to e.g. “the name of the thing we’re currently talking about as we evaluate this function”.
This all works with mutate(), too
# A tibble: 238 × 7
country world opt consent_law consent_practice consistent ccode
<chr> <chr> <chr> <chr> <chr> <chr> <chr>
1 AUSTRALIA LIBERAL IN INFORMED INFORMED YES OZ
2 AUSTRALIA LIBERAL IN INFORMED INFORMED YES OZ
3 AUSTRALIA LIBERAL IN INFORMED INFORMED YES OZ
4 AUSTRALIA LIBERAL IN INFORMED INFORMED YES OZ
5 AUSTRALIA LIBERAL IN INFORMED INFORMED YES OZ
6 AUSTRALIA LIBERAL IN INFORMED INFORMED YES OZ
7 AUSTRALIA LIBERAL IN INFORMED INFORMED YES OZ
8 AUSTRALIA LIBERAL IN INFORMED INFORMED YES OZ
9 AUSTRALIA LIBERAL IN INFORMED INFORMED YES OZ
10 AUSTRALIA LIBERAL IN INFORMED INFORMED YES OZ
# ℹ 228 more rowsArrange rows and columns
Sort rows with arrange()
organdata |>
group_by(consent_law, country) |>
summarize(donors = mean(donors, na.rm = TRUE)) |>
arrange(donors) |> ##<
print(n = 5)# A tibble: 17 × 3
# Groups: consent_law [2]
consent_law country donors
<chr> <chr> <dbl>
1 Informed Australia 10.6
2 Presumed Italy 11.1
3 Informed Germany 13.0
4 Informed Denmark 13.1
5 Presumed Sweden 13.1
# ℹ 12 more rowsArrange rows and columns
Sort rows with arrange()
organdata |>
group_by(consent_law, country) |>
summarize(donors = mean(donors, na.rm = TRUE)) |>
arrange(donors) |> ##<
print(n = 5)# A tibble: 17 × 3
# Groups: consent_law [2]
consent_law country donors
<chr> <chr> <dbl>
1 Informed Australia 10.6
2 Presumed Italy 11.1
3 Informed Germany 13.0
4 Informed Denmark 13.1
5 Presumed Sweden 13.1
# ℹ 12 more rowsorgandata |>
group_by(consent_law, country) |>
summarize(donors = mean(donors, na.rm = TRUE)) |>
arrange(desc(donors)) |> ##<
print(n = 5)# A tibble: 17 × 3
# Groups: consent_law [2]
consent_law country donors
<chr> <chr> <dbl>
1 Presumed Spain 28.1
2 Presumed Austria 23.5
3 Presumed Belgium 21.9
4 Informed United States 20.0
5 Informed Ireland 19.8
# ℹ 12 more rowsUsing arrange() to order rows in this way won’t respect groupings.
More generally …
organdata |>
group_by(consent_law, country) |>
summarize(donors = mean(donors, na.rm = TRUE)) |>
slice_max(donors, n = 5) # A tibble: 10 × 3
# Groups: consent_law [2]
consent_law country donors
<chr> <chr> <dbl>
1 Informed United States 20.0
2 Informed Ireland 19.8
3 Informed Canada 14.0
4 Informed Netherlands 13.7
5 Informed United Kingdom 13.5
6 Presumed Spain 28.1
7 Presumed Austria 23.5
8 Presumed Belgium 21.9
9 Presumed Finland 18.4
10 Presumed France 16.8- You can see that
slice_max()respects grouping. - There’s
slice_min(),slice_head(),slice_tail(),slice_sample(), and the most general one,slice().
dplyr’s window functions
Ranking and cumulation within groups.
# A tibble: 4,966 × 11
date year_week cname iso3 pop cases deaths cu_cases cu_deaths
<date> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 2019-12-30 2020-01 Austria AUT 8932664 NA NA NA NA
2 2020-01-06 2020-02 Austria AUT 8932664 NA NA NA NA
3 2020-01-13 2020-03 Austria AUT 8932664 NA NA NA NA
4 2020-01-20 2020-04 Austria AUT 8932664 NA NA NA NA
5 2020-01-27 2020-05 Austria AUT 8932664 NA NA NA NA
6 2020-02-03 2020-06 Austria AUT 8932664 NA NA NA NA
7 2020-02-10 2020-07 Austria AUT 8932664 NA NA NA NA
8 2020-02-17 2020-08 Austria AUT 8932664 NA NA NA NA
9 2020-02-24 2020-09 Austria AUT 8932664 12 0 12 0
10 2020-03-02 2020-10 Austria AUT 8932664 115 0 127 0
# ℹ 4,956 more rows
# ℹ 2 more variables: r14_cases <dbl>, r14_deaths <dbl>dplyr’s window functions
cumsum() gives cumulative sums
covnat_weekly |>
filter(iso3 == "FRA") |>
select(date, cname, iso3, cases) |>
mutate(cases = ifelse(is.na(cases), 0, cases), # convert NA vals in `cases` to 0
cumulative = cumsum(cases)) # A tibble: 159 × 5
date cname iso3 cases cumulative
<date> <chr> <chr> <dbl> <dbl>
1 2019-12-30 France FRA 0 0
2 2020-01-06 France FRA 0 0
3 2020-01-13 France FRA 0 0
4 2020-01-20 France FRA 3 3
5 2020-01-27 France FRA 3 6
6 2020-02-03 France FRA 6 12
7 2020-02-10 France FRA 0 12
8 2020-02-17 France FRA 4 16
9 2020-02-24 France FRA 133 149
10 2020-03-02 France FRA 981 1130
# ℹ 149 more rowsdplyr’s window functions
cume_dist() gives the proportion of values <= to the current value.
covnat_weekly |>
select(date, cname, iso3, deaths) |>
filter(iso3 == "FRA") |>
filter(cume_dist(desc(deaths)) < 0.1) # i.e. Top 10%# A tibble: 15 × 4
date cname iso3 deaths
<date> <chr> <chr> <dbl>
1 2020-04-06 France FRA 3348
2 2020-10-26 France FRA 3517
3 2020-11-02 France FRA 5281
4 2020-11-09 France FRA 6018
5 2020-11-16 France FRA 6208
6 2020-11-23 France FRA 5215
7 2020-11-30 France FRA 4450
8 2020-12-07 France FRA 4257
9 2020-12-14 France FRA 3786
10 2020-12-21 France FRA 3560
11 2021-01-04 France FRA 3851
12 2021-01-11 France FRA 3833
13 2021-01-18 France FRA 3754
14 2021-01-25 France FRA 3535
15 2021-02-01 France FRA 3431An application
covus |>
filter(measure == "death") |>
group_by(state) |>
arrange(state, desc(date)) |>
filter(state %in% "NY")# A tibble: 371 × 7
# Groups: state [1]
date state fips data_quality_grade measure count measure_label
<date> <chr> <chr> <lgl> <chr> <dbl> <chr>
1 2021-03-07 NY 36 NA death 39029 Deaths
2 2021-03-06 NY 36 NA death 38970 Deaths
3 2021-03-05 NY 36 NA death 38891 Deaths
4 2021-03-04 NY 36 NA death 38796 Deaths
5 2021-03-03 NY 36 NA death 38735 Deaths
6 2021-03-02 NY 36 NA death 38660 Deaths
7 2021-03-01 NY 36 NA death 38577 Deaths
8 2021-02-28 NY 36 NA death 38497 Deaths
9 2021-02-27 NY 36 NA death 38407 Deaths
10 2021-02-26 NY 36 NA death 38321 Deaths
# ℹ 361 more rowsHere the count measure is cumulative deaths. What if we want to recover the daily count for all the states in the data?
An application
dplyr has lead() and lag() functions. These allow you to access the previous and next values in a vector. You can calculate offsets this way.
An application
We can write the expression directly:
covus |>
select(-data_quality_grade) |>
filter(measure == "death") |>
group_by(state) |>
arrange(date) |>
mutate(deaths_daily = count - lag(count, order_by = date)) |>
arrange(state, desc(date)) |>
filter(state %in% "NY")# A tibble: 371 × 7
# Groups: state [1]
date state fips measure count measure_label deaths_daily
<date> <chr> <chr> <chr> <dbl> <chr> <dbl>
1 2021-03-07 NY 36 death 39029 Deaths 59
2 2021-03-06 NY 36 death 38970 Deaths 79
3 2021-03-05 NY 36 death 38891 Deaths 95
4 2021-03-04 NY 36 death 38796 Deaths 61
5 2021-03-03 NY 36 death 38735 Deaths 75
6 2021-03-02 NY 36 death 38660 Deaths 83
7 2021-03-01 NY 36 death 38577 Deaths 80
8 2021-02-28 NY 36 death 38497 Deaths 90
9 2021-02-27 NY 36 death 38407 Deaths 86
10 2021-02-26 NY 36 death 38321 Deaths 94
# ℹ 361 more rowsWriting our own functions
We write functions using the special function() function.*
Writing our own functions
We write our function. It’s just the expression we originally wrote, wrapped up.
This function has no generality, error-handling, or anything else. It’s a once-off.
Writing our own functions
Now we can use it like any other:
covus |>
filter(measure == "death") |>
select(-data_quality_grade) |>
group_by(state) |>
arrange(date) |>
mutate(deaths_daily = get_daily_count(count, date)) |>
arrange(state, desc(date)) |>
filter(state %in% "NY")# A tibble: 371 × 7
# Groups: state [1]
date state fips measure count measure_label deaths_daily
<date> <chr> <chr> <chr> <dbl> <chr> <dbl>
1 2021-03-07 NY 36 death 39029 Deaths 59
2 2021-03-06 NY 36 death 38970 Deaths 79
3 2021-03-05 NY 36 death 38891 Deaths 95
4 2021-03-04 NY 36 death 38796 Deaths 61
5 2021-03-03 NY 36 death 38735 Deaths 75
6 2021-03-02 NY 36 death 38660 Deaths 83
7 2021-03-01 NY 36 death 38577 Deaths 80
8 2021-02-28 NY 36 death 38497 Deaths 90
9 2021-02-27 NY 36 death 38407 Deaths 86
10 2021-02-26 NY 36 death 38321 Deaths 94
# ℹ 361 more rowsNot super-useful quite yet, but if our task had more steps …
The slider package
Tidy moving averages with slider
dplyr’s window functions don’t include moving averages.
There are several options, notably RcppRoll
We’ll use the slider package.
Tidy moving averages with slider
covus |>
filter(measure == "death") |>
select(-data_quality_grade) |>
group_by(state) |>
arrange(date) |>
mutate(
deaths_daily = get_daily_count(count, date),
deaths7 = slide_mean(deaths_daily,
before = 7,
na_rm = TRUE)) |>
arrange(state, desc(date)) |>
filter(state %in% "NY")# A tibble: 371 × 8
# Groups: state [1]
date state fips measure count measure_label deaths_daily deaths7
<date> <chr> <chr> <chr> <dbl> <chr> <dbl> <dbl>
1 2021-03-07 NY 36 death 39029 Deaths 59 77.8
2 2021-03-06 NY 36 death 38970 Deaths 79 81.1
3 2021-03-05 NY 36 death 38891 Deaths 95 83
4 2021-03-04 NY 36 death 38796 Deaths 61 82.6
5 2021-03-03 NY 36 death 38735 Deaths 75 88
6 2021-03-02 NY 36 death 38660 Deaths 83 89.9
7 2021-03-01 NY 36 death 38577 Deaths 80 90.8
8 2021-02-28 NY 36 death 38497 Deaths 90 90.1
9 2021-02-27 NY 36 death 38407 Deaths 86 91.5
10 2021-02-26 NY 36 death 38321 Deaths 94 95.6
# ℹ 361 more rowsTidy moving averages with slider
Notice the Tidyverse-style na_rm argument rather than the usual base na.rm
The package provides a lot of different functions, from general-purpose slide_max(), slide_min() to more specialized sliding functions. In particular note e.g. slide_index_mean() that addresses some subtleties in averaging over dates with gaps.
Move columns with relocate()
# A tibble: 2,867 × 32
year id ballot age childs sibs degree race sex region income16
<dbl> <dbl> <labelled> <dbl> <dbl> <labe> <fct> <fct> <fct> <fct> <fct>
1 2016 1 1 47 3 2 Bache… White Male New E… $170000…
2 2016 2 2 61 0 3 High … White Male New E… $50000 …
3 2016 3 3 72 2 3 Bache… White Male New E… $75000 …
4 2016 4 1 43 4 3 High … White Fema… New E… $170000…
5 2016 5 3 55 2 2 Gradu… White Fema… New E… $170000…
6 2016 6 2 53 2 2 Junio… White Fema… New E… $60000 …
7 2016 7 1 50 2 2 High … White Male New E… $170000…
8 2016 8 3 23 3 6 High … Other Fema… Middl… $30000 …
9 2016 9 1 45 3 5 High … Black Male Middl… $60000 …
10 2016 10 3 71 4 1 Junio… White Male Middl… $60000 …
# ℹ 2,857 more rows
# ℹ 21 more variables: relig <fct>, marital <fct>, padeg <fct>, madeg <fct>,
# partyid <fct>, polviews <fct>, happy <fct>, partners <fct>, grass <fct>,
# zodiac <fct>, pres12 <labelled>, wtssall <dbl>, income_rc <fct>,
# agegrp <fct>, ageq <fct>, siblings <fct>, kids <fct>, religion <fct>,
# bigregion <fct>, partners_rc <fct>, obama <dbl>Shuffle columns around
# A tibble: 2,867 × 32
year id ballot age childs sibs degree race sex region income16
<dbl> <dbl> <labelled> <dbl> <dbl> <labe> <fct> <fct> <fct> <fct> <fct>
1 2016 1 1 47 3 2 Bache… White Male New E… $170000…
2 2016 2 2 61 0 3 High … White Male New E… $50000 …
3 2016 3 3 72 2 3 Bache… White Male New E… $75000 …
4 2016 4 1 43 4 3 High … White Fema… New E… $170000…
5 2016 5 3 55 2 2 Gradu… White Fema… New E… $170000…
6 2016 6 2 53 2 2 Junio… White Fema… New E… $60000 …
7 2016 7 1 50 2 2 High … White Male New E… $170000…
8 2016 8 3 23 3 6 High … Other Fema… Middl… $30000 …
9 2016 9 1 45 3 5 High … Black Male Middl… $60000 …
10 2016 10 3 71 4 1 Junio… White Male Middl… $60000 …
# ℹ 2,857 more rows
# ℹ 21 more variables: relig <fct>, marital <fct>, padeg <fct>, madeg <fct>,
# partyid <fct>, polviews <fct>, happy <fct>, partners <fct>, grass <fct>,
# zodiac <fct>, pres12 <labelled>, wtssall <dbl>, income_rc <fct>,
# agegrp <fct>, ageq <fct>, siblings <fct>, kids <fct>, religion <fct>,
# bigregion <fct>, partners_rc <fct>, obama <dbl>Shuffle columns around
# A tibble: 2,867 × 19
region bigregion year id ballot age childs sibs degree race sex
<fct> <fct> <dbl> <dbl> <labe> <dbl> <dbl> <lab> <fct> <fct> <fct>
1 New Engla… Northeast 2016 1 1 47 3 2 Bache… White Male
2 New Engla… Northeast 2016 2 2 61 0 3 High … White Male
3 New Engla… Northeast 2016 3 3 72 2 3 Bache… White Male
4 New Engla… Northeast 2016 4 1 43 4 3 High … White Fema…
5 New Engla… Northeast 2016 5 3 55 2 2 Gradu… White Fema…
6 New Engla… Northeast 2016 6 2 53 2 2 Junio… White Fema…
7 New Engla… Northeast 2016 7 1 50 2 2 High … White Male
8 Middle At… Northeast 2016 8 3 23 3 6 High … Other Fema…
9 Middle At… Northeast 2016 9 1 45 3 5 High … Black Male
10 Middle At… Northeast 2016 10 3 71 4 1 Junio… White Male
# ℹ 2,857 more rows
# ℹ 8 more variables: padeg <fct>, partyid <fct>, polviews <fct>,
# partners <fct>, pres12 <labelled>, partners_rc <fct>, income16 <fct>,
# income_rc <fct>Shuffle columns around
# A tibble: 2,867 × 19
region bigregion year id ballot age children siblings degree race
<fct> <fct> <dbl> <dbl> <labe> <dbl> <dbl> <labell> <fct> <fct>
1 New England Northeast 2016 1 1 47 3 2 Bache… White
2 New England Northeast 2016 2 2 61 0 3 High … White
3 New England Northeast 2016 3 3 72 2 3 Bache… White
4 New England Northeast 2016 4 1 43 4 3 High … White
5 New England Northeast 2016 5 3 55 2 2 Gradu… White
6 New England Northeast 2016 6 2 53 2 2 Junio… White
7 New England Northeast 2016 7 1 50 2 2 High … White
8 Middle Atl… Northeast 2016 8 3 23 3 6 High … Other
9 Middle Atl… Northeast 2016 9 1 45 3 5 High … Black
10 Middle Atl… Northeast 2016 10 3 71 4 1 Junio… White
# ℹ 2,857 more rows
# ℹ 9 more variables: sex <fct>, padeg <fct>, partyid <fct>, polviews <fct>,
# partners <fct>, pres12 <labelled>, partners_rc <fct>, income16 <fct>,
# income_rc <fct>Shuffle columns around
# A tibble: 2,867 × 19
id region bigregion year ballot age children siblings degree race
<dbl> <fct> <fct> <dbl> <labe> <dbl> <dbl> <labell> <fct> <fct>
1 1 New England Northeast 2016 1 47 3 2 Bache… White
2 2 New England Northeast 2016 2 61 0 3 High … White
3 3 New England Northeast 2016 3 72 2 3 Bache… White
4 4 New England Northeast 2016 1 43 4 3 High … White
5 5 New England Northeast 2016 3 55 2 2 Gradu… White
6 6 New England Northeast 2016 2 53 2 2 Junio… White
7 7 New England Northeast 2016 1 50 2 2 High … White
8 8 Middle Atl… Northeast 2016 3 23 3 6 High … Other
9 9 Middle Atl… Northeast 2016 1 45 3 5 High … Black
10 10 Middle Atl… Northeast 2016 3 71 4 1 Junio… White
# ℹ 2,857 more rows
# ℹ 9 more variables: sex <fct>, padeg <fct>, partyid <fct>, polviews <fct>,
# partners <fct>, pres12 <labelled>, partners_rc <fct>, income16 <fct>,
# income_rc <fct>Shuffle columns around
# A tibble: 2,867 × 18
id region bigregion year age children siblings degree race sex padeg
<dbl> <fct> <fct> <dbl> <dbl> <dbl> <labell> <fct> <fct> <fct> <fct>
1 1 New E… Northeast 2016 47 3 2 Bache… White Male Grad…
2 2 New E… Northeast 2016 61 0 3 High … White Male Lt H…
3 3 New E… Northeast 2016 72 2 3 Bache… White Male High…
4 4 New E… Northeast 2016 43 4 3 High … White Fema… <NA>
5 5 New E… Northeast 2016 55 2 2 Gradu… White Fema… Bach…
6 6 New E… Northeast 2016 53 2 2 Junio… White Fema… <NA>
7 7 New E… Northeast 2016 50 2 2 High … White Male High…
8 8 Middl… Northeast 2016 23 3 6 High … Other Fema… Lt H…
9 9 Middl… Northeast 2016 45 3 5 High … Black Male Lt H…
10 10 Middl… Northeast 2016 71 4 1 Junio… White Male High…
# ℹ 2,857 more rows
# ℹ 7 more variables: partyid <fct>, polviews <fct>, partners <fct>,
# pres12 <labelled>, partners_rc <fct>, income16 <fct>, income_rc <fct>Shuffle columns around
# A tibble: 2,867 × 18
id year age children siblings pres12 region bigregion degree race
<dbl> <dbl> <dbl> <dbl> <labelled> <labelle> <fct> <fct> <fct> <fct>
1 1 2016 47 3 2 3 New E… Northeast Bache… White
2 2 2016 61 0 3 1 New E… Northeast High … White
3 3 2016 72 2 3 2 New E… Northeast Bache… White
4 4 2016 43 4 3 2 New E… Northeast High … White
5 5 2016 55 2 2 1 New E… Northeast Gradu… White
6 6 2016 53 2 2 1 New E… Northeast Junio… White
7 7 2016 50 2 2 NA New E… Northeast High … White
8 8 2016 23 3 6 NA Middl… Northeast High … Other
9 9 2016 45 3 5 NA Middl… Northeast High … Black
10 10 2016 71 4 1 2 Middl… Northeast Junio… White
# ℹ 2,857 more rows
# ℹ 8 more variables: sex <fct>, padeg <fct>, partyid <fct>, polviews <fct>,
# partners <fct>, partners_rc <fct>, income16 <fct>, income_rc <fct>Shuffle columns around
# A tibble: 2,867 × 18
id year region bigregion age children siblings pres12 degree race
<dbl> <dbl> <fct> <fct> <dbl> <dbl> <labell> <labe> <fct> <fct>
1 1 2016 New England Northeast 47 3 2 3 Bache… White
2 2 2016 New England Northeast 61 0 3 1 High … White
3 3 2016 New England Northeast 72 2 3 2 Bache… White
4 4 2016 New England Northeast 43 4 3 2 High … White
5 5 2016 New England Northeast 55 2 2 1 Gradu… White
6 6 2016 New England Northeast 53 2 2 1 Junio… White
7 7 2016 New England Northeast 50 2 2 NA High … White
8 8 2016 Middle Atl… Northeast 23 3 6 NA High … Other
9 9 2016 Middle Atl… Northeast 45 3 5 NA High … Black
10 10 2016 Middle Atl… Northeast 71 4 1 2 Junio… White
# ℹ 2,857 more rows
# ℹ 8 more variables: sex <fct>, padeg <fct>, partyid <fct>, polviews <fct>,
# partners <fct>, partners_rc <fct>, income16 <fct>, income_rc <fct>Example: UK Election Data
# A tibble: 3,320 × 13
cid constituency electorate party_name candidate votes vote_share_percent
<chr> <chr> <int> <chr> <chr> <int> <dbl>
1 W07000… Aberavon 50747 Labour Stephen … 17008 53.8
2 W07000… Aberavon 50747 Conservat… Charlott… 6518 20.6
3 W07000… Aberavon 50747 The Brexi… Glenda D… 3108 9.8
4 W07000… Aberavon 50747 Plaid Cym… Nigel Hu… 2711 8.6
5 W07000… Aberavon 50747 Liberal D… Sheila K… 1072 3.4
6 W07000… Aberavon 50747 Independe… Captain … 731 2.3
7 W07000… Aberavon 50747 Green Giorgia … 450 1.4
8 W07000… Aberconwy 44699 Conservat… Robin Mi… 14687 46.1
9 W07000… Aberconwy 44699 Labour Emily Ow… 12653 39.7
10 W07000… Aberconwy 44699 Plaid Cym… Lisa Goo… 2704 8.5
# ℹ 3,310 more rows
# ℹ 6 more variables: vote_share_change <dbl>, total_votes_cast <int>,
# vrank <int>, turnout <dbl>, fname <chr>, lname <chr>Example: UK Election Data
Use sample_n() to sample n rows of your tibble.
# A tibble: 10 × 13
cid constituency electorate party_name candidate votes vote_share_percent
<chr> <chr> <int> <chr> <chr> <int> <dbl>
1 E14000… Garston & H… 76116 Conservat… Neva Nov… 6954 13
2 E14000… Bradford We… 70694 Independe… Azfar Bu… 90 0.2
3 S14000… Ayrshire No… 73534 Conservat… David Ro… 14855 30.8
4 S14000… Fife North … 60905 Scottish … Stephen … 18447 40.2
5 E14000… Stalybridge… 73873 Labour Jonathan… 19025 44.9
6 E14000… Basildon & … 69906 Liberal D… Edward S… 3741 8.5
7 E14000… Gainsborough 76343 Labour Perry Sm… 10926 21.4
8 W07000… Monmouth 67094 Plaid Cym… Hugh Koc… 1182 2.4
9 E14000… Bournemouth… 74211 Conservat… Conor Bu… 24550 53.4
10 E14000… Calder Vall… 79287 Liberal D… Javed Ba… 2884 5
# ℹ 6 more variables: vote_share_change <dbl>, total_votes_cast <int>,
# vrank <int>, turnout <dbl>, fname <chr>, lname <chr>Example: UK Election Data
- A vector of unique constituency names
Example: UK Election Data
- Tally them up
Example: UK Election Data
Which parties fielded the most candidates?
# A tibble: 69 × 2
party_name n
<chr> <int>
1 Conservative 636
2 Labour 631
3 Liberal Democrat 611
4 Green 497
5 The Brexit Party 275
6 Independent 224
7 Scottish National Party 59
8 UKIP 44
9 Plaid Cymru 36
10 Christian Peoples Alliance 29
# ℹ 59 more rowsExample: UK Election Data
Example: UK Election Data
- Top 5
- Bottom 5
# A tibble: 25 × 2
party_name n
<chr> <int>
1 Ashfield Independents 1
2 Best for Luton 1
3 Birkenhead Social Justice Party 1
4 British National Party 1
5 Burnley & Padiham Independent Party 1
6 Church of the Militant Elvis Party 1
7 Citizens Movement Party UK 1
8 CumbriaFirst 1
9 Heavy Woollen District Independents 1
10 Independent Network 1
# ℹ 15 more rowsExample: UK Election Data
How many constituencies are there?
# A tibble: 650 × 2
constituency n
<chr> <int>
1 Aberavon 7
2 Aberconwy 4
3 Aberdeen North 6
4 Aberdeen South 4
5 Aberdeenshire West & Kincardine 4
6 Airdrie & Shotts 5
7 Aldershot 4
8 Aldridge-Brownhills 5
9 Altrincham & Sale West 6
10 Alyn & Deeside 5
# ℹ 640 more rowsCounting Twice Over
Counting Twice Over
# A tibble: 3,320 × 13
cid constituency electorate party_name candidate votes vote_share_percent
<chr> <chr> <int> <chr> <chr> <int> <dbl>
1 W07000… Aberavon 50747 Labour Stephen … 17008 53.8
2 W07000… Aberavon 50747 Conservat… Charlott… 6518 20.6
3 W07000… Aberavon 50747 The Brexi… Glenda D… 3108 9.8
4 W07000… Aberavon 50747 Plaid Cym… Nigel Hu… 2711 8.6
5 W07000… Aberavon 50747 Liberal D… Sheila K… 1072 3.4
6 W07000… Aberavon 50747 Independe… Captain … 731 2.3
7 W07000… Aberavon 50747 Green Giorgia … 450 1.4
8 W07000… Aberconwy 44699 Conservat… Robin Mi… 14687 46.1
9 W07000… Aberconwy 44699 Labour Emily Ow… 12653 39.7
10 W07000… Aberconwy 44699 Plaid Cym… Lisa Goo… 2704 8.5
# ℹ 3,310 more rows
# ℹ 6 more variables: vote_share_change <dbl>, total_votes_cast <int>,
# vrank <int>, turnout <dbl>, fname <chr>, lname <chr>Counting Twice Over
# A tibble: 650 × 2
constituency n_cands
<chr> <int>
1 Aberavon 7
2 Aberconwy 4
3 Aberdeen North 6
4 Aberdeen South 4
5 Aberdeenshire West & Kincardine 4
6 Airdrie & Shotts 5
7 Aldershot 4
8 Aldridge-Brownhills 5
9 Altrincham & Sale West 6
10 Alyn & Deeside 5
# ℹ 640 more rowsCounting Twice Over
Recap and Looking Ahead
Recap and Looking Ahead
Coding as gardening
Working in RStudio with RMarkdown documents
Core dplyr verbs
- Subset your table:
filter()rows,select()columns - Logically
group_by()one or more columns - Add columns with
mutate() - Summarize (by group, or the whole table) with
summarize()
Expand your dplyr actions
- Count up rows with
n(),tally()orcount() - Calculate quantities with
sum(),mean(),min(), etc - Subset rows with logical expressions or
slicefunctions - Conditionally select columns by name directly, with
%in%or%nin%, or with tidy selectors likestarts_with(),ends_with(),contains() - Conditionally select columns by type with
where()and some criterion, e.g.where(is.numeric) - Conditionally select and then act on columns with
across(where(<condition>),<action>)
Expand your dplyr actions
- Tidy up columns with
relocate()andrename() - Tidy up rows with
arrange()
A dplyr shortcut
A dplyr shortcut
So far we have been writing, e.g.,
# A tibble: 24 × 3
# Groups: bigregion [4]
bigregion religion total
<fct> <fct> <int>
1 Northeast Protestant 158
2 Northeast Catholic 162
3 Northeast Jewish 27
4 Northeast None 112
5 Northeast Other 28
6 Northeast <NA> 1
7 Midwest Protestant 325
8 Midwest Catholic 172
9 Midwest Jewish 3
10 Midwest None 157
# ℹ 14 more rowsA dplyr shortcut
Or
# A tibble: 24 × 3
# Groups: bigregion [4]
bigregion religion n
<fct> <fct> <int>
1 Northeast Protestant 158
2 Northeast Catholic 162
3 Northeast Jewish 27
4 Northeast None 112
5 Northeast Other 28
6 Northeast <NA> 1
7 Midwest Protestant 325
8 Midwest Catholic 172
9 Midwest Jewish 3
10 Midwest None 157
# ℹ 14 more rowsA dplyr shortcut
Or
# A tibble: 24 × 3
bigregion religion n
<fct> <fct> <int>
1 Northeast Protestant 158
2 Northeast Catholic 162
3 Northeast Jewish 27
4 Northeast None 112
5 Northeast Other 28
6 Northeast <NA> 1
7 Midwest Protestant 325
8 Midwest Catholic 172
9 Midwest Jewish 3
10 Midwest None 157
# ℹ 14 more rowsWith this last one the final result is ungrouped, no matter how many levels of grouping there are going in.
A dplyr shortcut
But we can also write this:
# A tibble: 24 × 3
bigregion religion total
<fct> <fct> <int>
1 Northeast None 112
2 Northeast Catholic 162
3 Northeast Protestant 158
4 Northeast Other 28
5 Northeast Jewish 27
6 West Jewish 10
7 West None 180
8 West Other 48
9 West Protestant 238
10 West Catholic 155
# ℹ 14 more rowsBy default the result is an ungrouped tibble, whereas with group_by() … summarize() the result would still be grouped by bigregion at the end. To prevent unexpected results, you can’t use .by on tibble that’s already grouped.
Data as implicitly first
This code:
# A tibble: 24 × 3
bigregion religion total
<fct> <fct> <int>
1 Northeast None 112
2 Northeast Catholic 162
3 Northeast Protestant 158
4 Northeast Other 28
5 Northeast Jewish 27
6 West Jewish 10
7 West None 180
8 West Other 48
9 West Protestant 238
10 West Catholic 155
# ℹ 14 more rowsData as implicitly first
… is equivalent to this:
# A tibble: 24 × 3
bigregion religion total
<fct> <fct> <int>
1 Northeast None 112
2 Northeast Catholic 162
3 Northeast Protestant 158
4 Northeast Other 28
5 Northeast Jewish 27
6 West Jewish 10
7 West None 180
8 West Other 48
9 West Protestant 238
10 West Catholic 155
# ℹ 14 more rowsThis is true of Tidyverse pipelines in general. Let’s look at the help for summarize() to see why.
Two dplyr gotchas
Comparisons filtering on proportions
Let’s say you are working with proportions …
Comparisons filtering on proportions
And you want to focus on cases where prop1 plus prop2 is greater than 0.3:
# A tibble: 3 × 3
id prop1 prop2
<chr> <dbl> <dbl>
1 A 0.1 0.2
2 B 0.1 0.21
3 C 0.11 0.2 - The row with
idAshouldn’t have been included there.
- This is not dplyr’s fault. It’s our floating point friend again.
Comparisons filtering on proportions
# A tibble: 0 × 3
# ℹ 3 variables: id <chr>, prop1 <dbl>, prop2 <dbl>The row with id A should have been included here!
Comparisons filtering on proportions
This won’t give the right behavior either:
Comparisons filtering on proportions
So, beware.
# A tibble: 1 × 3
id prop1 prop2
<chr> <dbl> <dbl>
1 A 0.1 0.2Better:
Zero Counts in dplyr
# A tibble: 280 × 4
pid start_year party sex
<dbl> <date> <chr> <chr>
1 3160 2013-01-03 Republican M
2 3161 2013-01-03 Democrat F
3 3162 2013-01-03 Democrat M
4 3163 2013-01-03 Republican M
5 3164 2013-01-03 Democrat M
6 3165 2013-01-03 Republican M
7 3166 2013-01-03 Republican M
8 3167 2013-01-03 Democrat F
9 3168 2013-01-03 Republican M
10 3169 2013-01-03 Democrat M
# ℹ 270 more rowsZero Counts in dplyr
# A tibble: 14 × 5
# Groups: start_year, party [8]
start_year party sex N freq
<date> <chr> <chr> <int> <dbl>
1 2013-01-03 Democrat F 21 0.362
2 2013-01-03 Democrat M 37 0.638
3 2013-01-03 Republican F 8 0.101
4 2013-01-03 Republican M 71 0.899
5 2015-01-03 Democrat M 1 1
6 2015-01-03 Republican M 5 1
7 2017-01-03 Democrat F 6 0.24
8 2017-01-03 Democrat M 19 0.76
9 2017-01-03 Republican F 2 0.0667
10 2017-01-03 Republican M 28 0.933
11 2019-01-03 Democrat F 33 0.647
12 2019-01-03 Democrat M 18 0.353
13 2019-01-03 Republican F 1 0.0323
14 2019-01-03 Republican M 30 0.968 Zero Counts in dplyr
p_col <- df |>
group_by(start_year, party, sex) |>
summarize(N = n()) |>
mutate(freq = N / sum(N)) |>
ggplot(aes(x = start_year,
y = freq,
fill = sex)) +
geom_col() +
scale_y_continuous(labels = scales::percent) +
scale_fill_manual(values = sex_colors, labels = c("Women", "Men")) +
labs(x = "Year", y = "Percent", fill = "Group") +
facet_wrap(~ party)Zero Counts in dplyr

2. Zero Counts in dplyr
p_line <- df |>
group_by(start_year, party, sex) |>
summarize(N = n()) |>
mutate(freq = N / sum(N)) |>
ggplot(aes(x = start_year,
y = freq,
color = sex)) +
geom_line(size = 1.1) +
scale_y_continuous(labels = scales::percent) +
scale_color_manual(values = sex_colors, labels = c("Women", "Men")) +
guides(color = guide_legend(reverse = TRUE)) +
labs(x = "Year", y = "Percent", color = "Group") +
facet_wrap(~ party)Zero Counts in dplyr

Option 1: factors and .drop
Factors are for categorical variables and are stored differently from characters.
This can matter when modeling, and also now.
# A tibble: 280 × 5
pid start_year party sex party_f
<dbl> <date> <chr> <chr> <fct>
1 3160 2013-01-03 Republican M Republican
2 3161 2013-01-03 Democrat F Democrat
3 3162 2013-01-03 Democrat M Democrat
4 3163 2013-01-03 Republican M Republican
5 3164 2013-01-03 Democrat M Democrat
6 3165 2013-01-03 Republican M Republican
7 3166 2013-01-03 Republican M Republican
8 3167 2013-01-03 Democrat F Democrat
9 3168 2013-01-03 Republican M Republican
10 3169 2013-01-03 Democrat M Democrat
# ℹ 270 more rowsOption 1: factors and .drop
# A tibble: 2 × 2
party_f n
<fct> <int>
1 Democrat 135
2 Republican 145Factors are integer values with named labels, or levels:
Option 1: factors and .drop
By default, unused levels won’t display:
Option 1: factors and .drop
By default, unused levels won’t display:
df |>
mutate(across(where(is.character), as_factor)) |>
group_by(start_year, party, sex) |>
summarize(N = n()) |>
mutate(freq = N / sum(N))# A tibble: 14 × 5
# Groups: start_year, party [8]
start_year party sex N freq
<date> <fct> <fct> <int> <dbl>
1 2013-01-03 Republican M 71 0.899
2 2013-01-03 Republican F 8 0.101
3 2013-01-03 Democrat M 37 0.638
4 2013-01-03 Democrat F 21 0.362
5 2015-01-03 Republican M 5 1
6 2015-01-03 Democrat M 1 1
7 2017-01-03 Republican M 28 0.933
8 2017-01-03 Republican F 2 0.0667
9 2017-01-03 Democrat M 19 0.76
10 2017-01-03 Democrat F 6 0.24
11 2019-01-03 Republican M 30 0.968
12 2019-01-03 Republican F 1 0.0323
13 2019-01-03 Democrat M 18 0.353
14 2019-01-03 Democrat F 33 0.647 Option 1: factors and .drop
You can make dplyr keep empty factor levels though:
df |>
mutate(across(where(is.character), as_factor)) |>
group_by(start_year, party, sex, .drop = FALSE) |>
summarize(N = n()) |>
mutate(freq = N / sum(N))# A tibble: 16 × 5
# Groups: start_year, party [8]
start_year party sex N freq
<date> <fct> <fct> <int> <dbl>
1 2013-01-03 Republican M 71 0.899
2 2013-01-03 Republican F 8 0.101
3 2013-01-03 Democrat M 37 0.638
4 2013-01-03 Democrat F 21 0.362
5 2015-01-03 Republican M 5 1
6 2015-01-03 Republican F 0 0
7 2015-01-03 Democrat M 1 1
8 2015-01-03 Democrat F 0 0
9 2017-01-03 Republican M 28 0.933
10 2017-01-03 Republican F 2 0.0667
11 2017-01-03 Democrat M 19 0.76
12 2017-01-03 Democrat F 6 0.24
13 2019-01-03 Republican M 30 0.968
14 2019-01-03 Republican F 1 0.0323
15 2019-01-03 Democrat M 18 0.353
16 2019-01-03 Democrat F 33 0.647 Option 2: ungroup() and complete()
Maybe you don’t want to deal with factors.
Option 2: ungroup() and complete()
# A tibble: 16 × 5
start_year party sex N freq
<date> <chr> <chr> <int> <dbl>
1 2013-01-03 Democrat F 21 0.362
2 2013-01-03 Democrat M 37 0.638
3 2013-01-03 Republican F 8 0.101
4 2013-01-03 Republican M 71 0.899
5 2015-01-03 Democrat F 0 0
6 2015-01-03 Democrat M 1 1
7 2015-01-03 Republican F 0 0
8 2015-01-03 Republican M 5 1
9 2017-01-03 Democrat F 6 0.24
10 2017-01-03 Democrat M 19 0.76
11 2017-01-03 Republican F 2 0.0667
12 2017-01-03 Republican M 28 0.933
13 2019-01-03 Democrat F 33 0.647
14 2019-01-03 Democrat M 18 0.353
15 2019-01-03 Republican F 1 0.0323
16 2019-01-03 Republican M 30 0.968 Option 2: ungroup() and complete()
p_out <- df_c |>
ggplot(aes(x = start_year,
y = freq,
color = sex)) +
geom_line(size = 1.1) +
scale_y_continuous(labels = scales::percent) +
scale_color_manual(values = sex_colors, labels = c("Women", "Men")) +
guides(color = guide_legend(reverse = TRUE)) +
labs(x = "Year", y = "Percent", color = "Group") +
facet_wrap(~ party)Option 2: ungroup() and complete()
