library(here) # manage file paths
library(socviz) # data and some useful functions
library(tidyverse) # your friend and mine
library(haven) # for Stata, SAS, and SPSS filesIngesting Data
Modern Plain Text Social Science: Week 7
October 24, 2024
Reading in data with readr and haven
Load the packages, as always
We’ve put a lot of pieces in place at this point
Including several things we haven’t fully exploited yet
Data we want to get into R
- Nice, clean CSV files.
- More troublesome CSVs.
- Other plain-text formats.
- Foreign formats, like Stata.
- Quite messy things like tables on web pages.
- … and more besides.
Reading in CSV files
- CSV is not really a proper format at all!
- Base R has
read.csv() - Corresponding tidyverse “underscored” version:
read_csv(). - It is pickier and more talkative than the Base R version.
Where’s the data? Using here()
- If we’re loading a file, it’s coming from somewhere.
- If it’s on our local disk somewhere, we will need to interact with the file system. We should try to do this in a way that avoids absolute file paths.
Where’s the data? Using here()
- If we’re loading a file, it’s coming from somewhere.
- If it’s on our local disk somewhere, we will need to interact with the file system. We should try to do this in a way that avoids absolute file paths.
- We should also do it in a way that is platform independent.
- This makes it easier to share your work, move it around, etc. Projects should be self-contained.
Where’s the data? Using here()
- The
herepackage, andhere()function builds paths relative to the top level of your R project.
Where’s the data? Using here()
- This seminar’s files all live in an RStudio project. It looks like this:
/Users/kjhealy/Documents/courses/mptc
├── R
├── README.md
├── README.qmd
├── README_files
├── _extensions
├── _freeze
├── _quarto.yml
├── _site
├── _targets
├── _targets.R
├── _variables.yml
├── about
├── assets
├── assignment
├── content
├── data
├── deploy.sh
├── example
├── files
├── html
├── index.html
├── index.qmd
├── mptc.Rproj
├── renv
├── renv.lock
├── renv.lock.orig
├── schedule
├── site_libs
├── slides
├── staging
└── syllabusI want to load files from the files/examples/ folder, but I also want you to be able to load them. I’m writing this from somewhere deep in the slides folder, but you won’t be there. Also, I’m on a Mac, but you may not be.
Where’s the data? Using here()
So:
# A tibble: 238 × 21
country year donors pop pop.dens gdp gdp.lag health health.lag pubhealth
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Austra… NA NA 17065 0.220 16774 16591 1300 1224 4.8
2 Austra… 1991 12.1 17284 0.223 17171 16774 1379 1300 5.4
3 Austra… 1992 12.4 17495 0.226 17914 17171 1455 1379 5.4
4 Austra… 1993 12.5 17667 0.228 18883 17914 1540 1455 5.4
5 Austra… 1994 10.2 17855 0.231 19849 18883 1626 1540 5.4
6 Austra… 1995 10.2 18072 0.233 21079 19849 1737 1626 5.5
7 Austra… 1996 10.6 18311 0.237 21923 21079 1846 1737 5.6
8 Austra… 1997 10.3 18518 0.239 22961 21923 1948 1846 5.7
9 Austra… 1998 10.5 18711 0.242 24148 22961 2077 1948 5.9
10 Austra… 1999 8.67 18926 0.244 25445 24148 2231 2077 6.1
# ℹ 228 more rows
# ℹ 11 more variables: roads <dbl>, cerebvas <dbl>, assault <dbl>,
# external <dbl>, txp.pop <dbl>, world <chr>, opt <chr>, consent.law <chr>,
# consent.practice <chr>, consistent <chr>, ccode <chr>read_csv() comes in different varieties
read_csv()Field separator is a comma: ,
read_csv2()Field separator is a semicolon: ;
- Both are special cases of
read_delim()
Other species are also catered to
read_tsv()Tab separated.read_fwf()Fixed-width files.read_log()Log files (i.e. computer log files).read_lines()Just read in lines, without trying to parse them.read_table()Data that’s separated by one (or more) columns of space.
You can read files remotely, too
- You can give all of these functions local files, or they can point to URLs.
- Compressed files will be automatically uncompressed.
- (Be careful what you download from remote locations!)
# A tibble: 238 × 21
country year donors pop pop.dens gdp gdp.lag health health.lag pubhealth
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Austra… NA NA 17065 0.220 16774 16591 1300 1224 4.8
2 Austra… 1991 12.1 17284 0.223 17171 16774 1379 1300 5.4
3 Austra… 1992 12.4 17495 0.226 17914 17171 1455 1379 5.4
4 Austra… 1993 12.5 17667 0.228 18883 17914 1540 1455 5.4
5 Austra… 1994 10.2 17855 0.231 19849 18883 1626 1540 5.4
6 Austra… 1995 10.2 18072 0.233 21079 19849 1737 1626 5.5
7 Austra… 1996 10.6 18311 0.237 21923 21079 1846 1737 5.6
8 Austra… 1997 10.3 18518 0.239 22961 21923 1948 1846 5.7
9 Austra… 1998 10.5 18711 0.242 24148 22961 2077 1948 5.9
10 Austra… 1999 8.67 18926 0.244 25445 24148 2231 2077 6.1
# ℹ 228 more rows
# ℹ 11 more variables: roads <dbl>, cerebvas <dbl>, assault <dbl>,
# external <dbl>, txp.pop <dbl>, world <chr>, opt <chr>, consent.law <chr>,
# consent.practice <chr>, consistent <chr>, ccode <chr>An example: read_table()
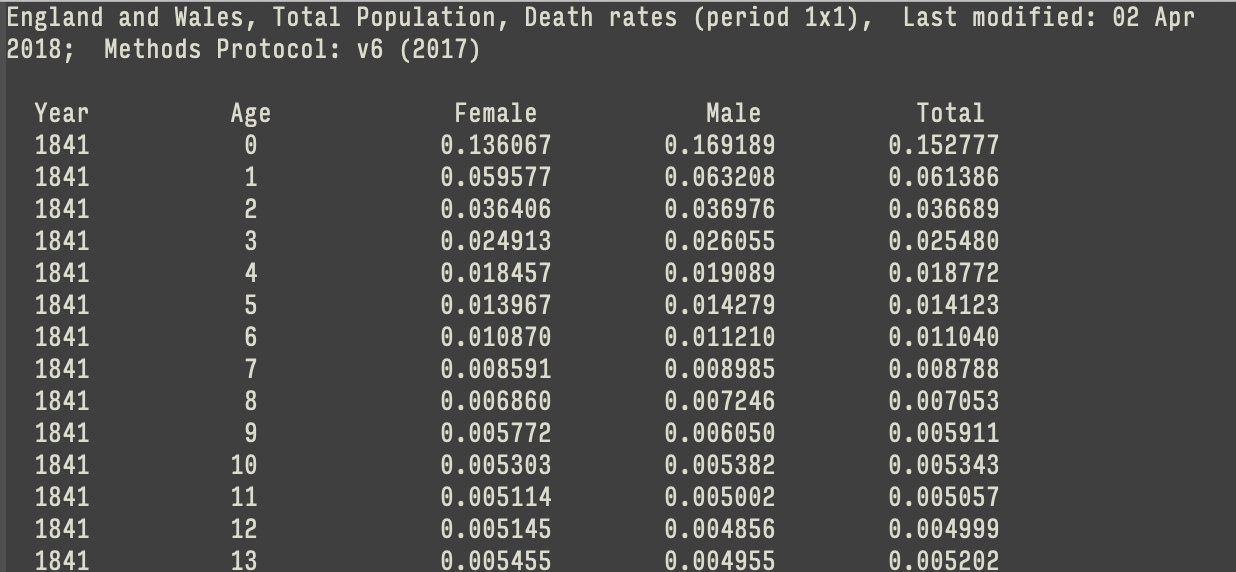
An example: read_table()


# A tibble: 222 × 5
Year Age Female Male Total
<dbl> <chr> <dbl> <dbl> <dbl>
1 1841 0 0.136 0.169 0.153
2 1841 1 0.0596 0.0632 0.0614
3 1841 2 0.0364 0.0370 0.0367
4 1841 3 0.0249 0.0261 0.0255
5 1841 4 0.0185 0.0191 0.0188
6 1841 5 0.0140 0.0143 0.0141
7 1841 6 0.0109 0.0112 0.0110
8 1841 7 0.00859 0.00898 0.00879
9 1841 8 0.00686 0.00725 0.00705
10 1841 9 0.00577 0.00605 0.00591
# ℹ 212 more rowsAttend to the column specification
── Column specification ────────────────────────────────────────────────────────
cols(
Year = col_double(),
Age = col_character(),
Female = col_double(),
Male = col_double(),
Total = col_double()
)- The column specification tells you what the read function did. That is, how it interpreted each of the columns. It will also report if things don’t go as expected.
- Why is
ageimported incharacterformat?
Attend to the column specification
Absent you giving them a column specification, the
read_functions try to guess what the type of each column is. They do this by looking at the first thousand rows of each column. They may guess incorrectly!For plain-text delimited formats like CSV, TSV, and the like, their
read_functions guess because they have to. Why? Because CSV is not really a format, and because CSV files do not carry metadata inside them about what their detailed structure is.
Normalizing names and recoding
# A tibble: 222 × 5
Year Age Female Male Total
<dbl> <chr> <dbl> <dbl> <dbl>
1 1841 0 0.136 0.169 0.153
2 1841 1 0.0596 0.0632 0.0614
3 1841 2 0.0364 0.0370 0.0367
4 1841 3 0.0249 0.0261 0.0255
5 1841 4 0.0185 0.0191 0.0188
6 1841 5 0.0140 0.0143 0.0141
7 1841 6 0.0109 0.0112 0.0110
8 1841 7 0.00859 0.00898 0.00879
9 1841 8 0.00686 0.00725 0.00705
10 1841 9 0.00577 0.00605 0.00591
# ℹ 212 more rowsNormalizing names and recoding
# A tibble: 222 × 5
year age female male total
<dbl> <chr> <dbl> <dbl> <dbl>
1 1841 0 0.136 0.169 0.153
2 1841 1 0.0596 0.0632 0.0614
3 1841 2 0.0364 0.0370 0.0367
4 1841 3 0.0249 0.0261 0.0255
5 1841 4 0.0185 0.0191 0.0188
6 1841 5 0.0140 0.0143 0.0141
7 1841 6 0.0109 0.0112 0.0110
8 1841 7 0.00859 0.00898 0.00879
9 1841 8 0.00686 0.00725 0.00705
10 1841 9 0.00577 0.00605 0.00591
# ℹ 212 more rowsNormalizing names and recoding
# A tibble: 222 × 5
year age female male total
<dbl> <int> <dbl> <dbl> <dbl>
1 1841 0 0.136 0.169 0.153
2 1841 1 0.0596 0.0632 0.0614
3 1841 2 0.0364 0.0370 0.0367
4 1841 3 0.0249 0.0261 0.0255
5 1841 4 0.0185 0.0191 0.0188
6 1841 5 0.0140 0.0143 0.0141
7 1841 6 0.0109 0.0112 0.0110
8 1841 7 0.00859 0.00898 0.00879
9 1841 8 0.00686 0.00725 0.00705
10 1841 9 0.00577 0.00605 0.00591
# ℹ 212 more rowsJanitor
- The
janitorpackage is very handy! - The main cost of normalizing names comes with, e.g., data where there is a codebook you need to consult. But in general it’s worth it.
Example: Colspecs
More on column specifications
- CDC/NCHS data: Provisional COVID-19 Death Counts by Sex, Age, and State
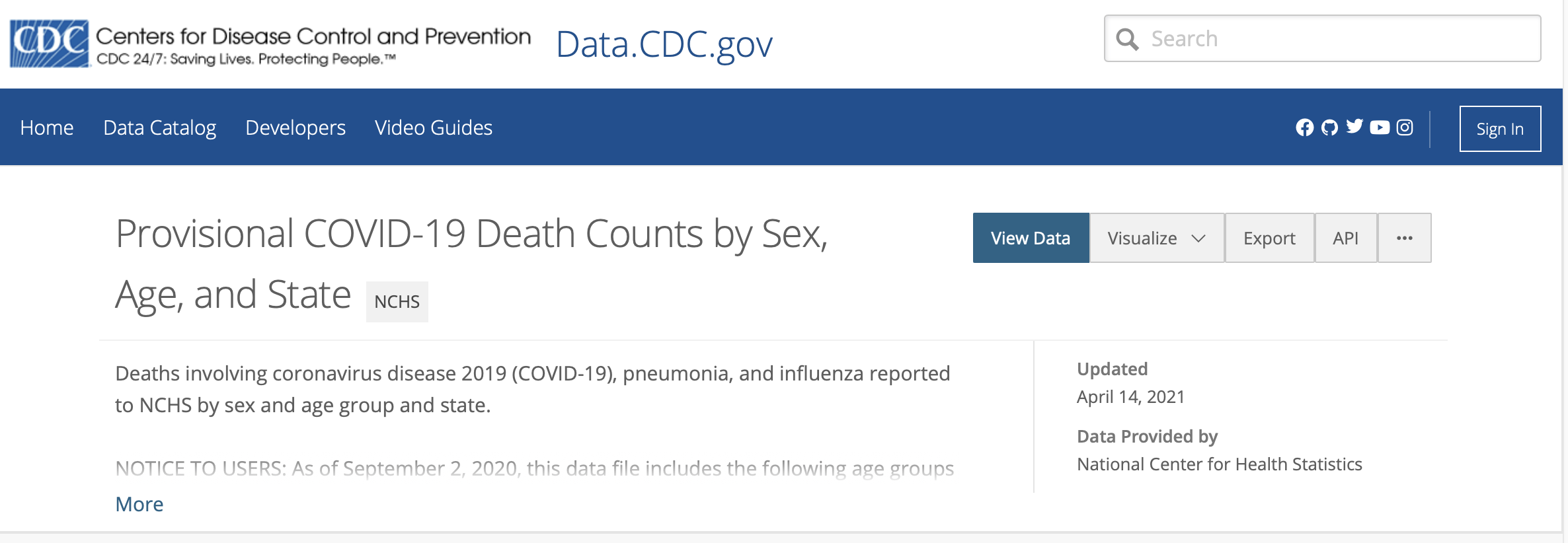
More on column specifications
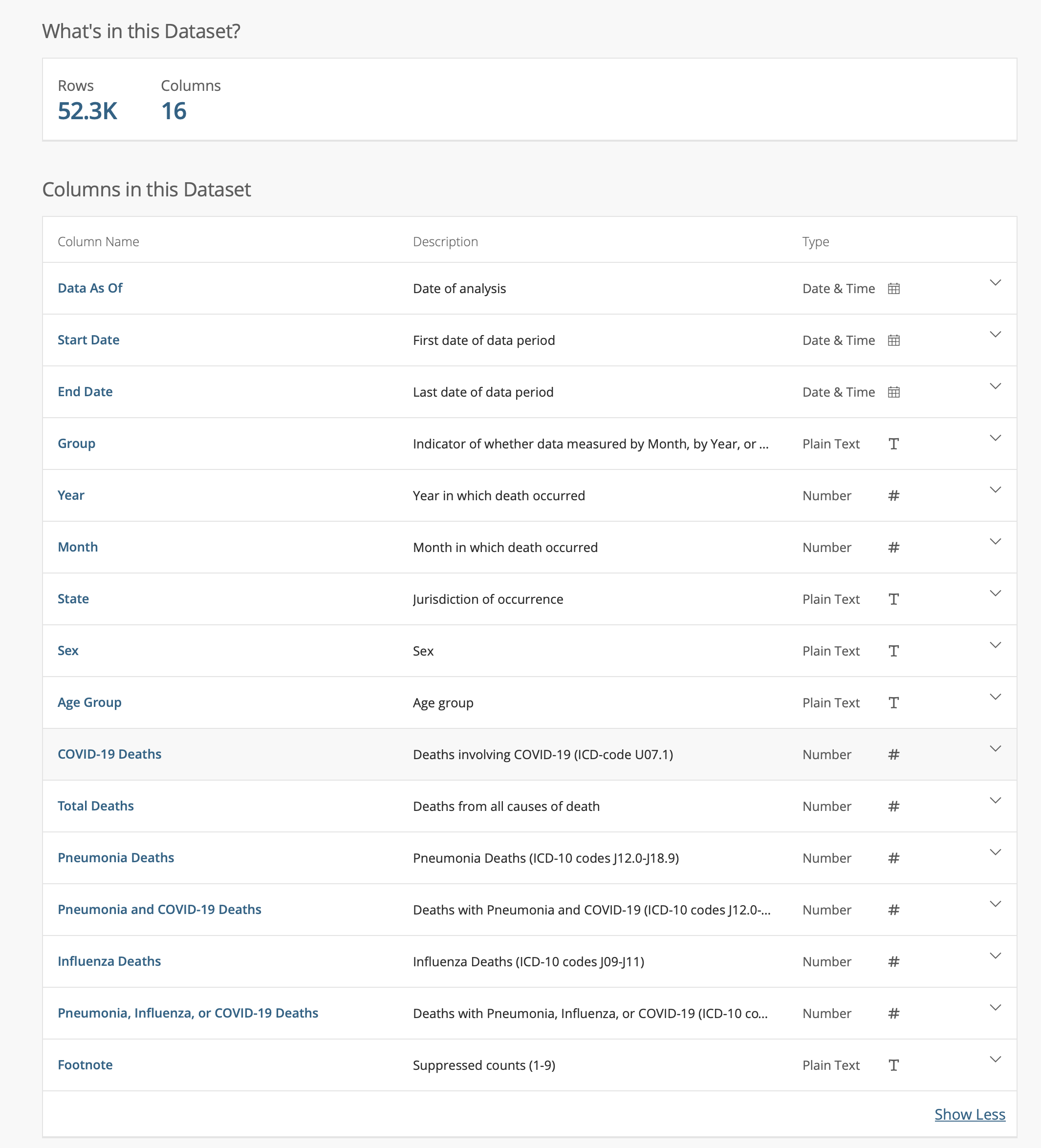
Let’s try to load it
── Column specification ────────────────────────────────────────────────────────
cols(
`Data As Of` = col_character(),
`Start Date` = col_character(),
`End Date` = col_character(),
Group = col_character(),
Year = col_logical(),
Month = col_logical(),
State = col_character(),
Sex = col_character(),
`Age Group` = col_character(),
`COVID-19 Deaths` = col_double(),
`Total Deaths` = col_double(),
`Pneumonia Deaths` = col_double(),
`Pneumonia and COVID-19 Deaths` = col_double(),
`Influenza Deaths` = col_double(),
`Pneumonia, Influenza, or COVID-19 Deaths` = col_double(),
Footnote = col_character()
)Warning: 88128 parsing failures.
row col expected actual file
2755 Year 1/0/T/F/TRUE/FALSE 2020 '/Users/kjhealy/Documents/courses/mptc/files/examples/SAS_on_2021-04-13.csv'
2756 Year 1/0/T/F/TRUE/FALSE 2020 '/Users/kjhealy/Documents/courses/mptc/files/examples/SAS_on_2021-04-13.csv'
2757 Year 1/0/T/F/TRUE/FALSE 2020 '/Users/kjhealy/Documents/courses/mptc/files/examples/SAS_on_2021-04-13.csv'
2758 Year 1/0/T/F/TRUE/FALSE 2020 '/Users/kjhealy/Documents/courses/mptc/files/examples/SAS_on_2021-04-13.csv'
2759 Year 1/0/T/F/TRUE/FALSE 2020 '/Users/kjhealy/Documents/courses/mptc/files/examples/SAS_on_2021-04-13.csv'
.... .... .................. ...... ............................................................................
See problems(...) for more details.Let’s try to load it
# A tibble: 88,128 × 5
row col expected actual file
<int> <chr> <chr> <chr> <chr>
1 2755 Year 1/0/T/F/TRUE/FALSE 2020 '/Users/kjhealy/Documents/courses/mptc…
2 2756 Year 1/0/T/F/TRUE/FALSE 2020 '/Users/kjhealy/Documents/courses/mptc…
3 2757 Year 1/0/T/F/TRUE/FALSE 2020 '/Users/kjhealy/Documents/courses/mptc…
4 2758 Year 1/0/T/F/TRUE/FALSE 2020 '/Users/kjhealy/Documents/courses/mptc…
5 2759 Year 1/0/T/F/TRUE/FALSE 2020 '/Users/kjhealy/Documents/courses/mptc…
6 2760 Year 1/0/T/F/TRUE/FALSE 2020 '/Users/kjhealy/Documents/courses/mptc…
7 2761 Year 1/0/T/F/TRUE/FALSE 2020 '/Users/kjhealy/Documents/courses/mptc…
8 2762 Year 1/0/T/F/TRUE/FALSE 2020 '/Users/kjhealy/Documents/courses/mptc…
9 2763 Year 1/0/T/F/TRUE/FALSE 2020 '/Users/kjhealy/Documents/courses/mptc…
10 2764 Year 1/0/T/F/TRUE/FALSE 2020 '/Users/kjhealy/Documents/courses/mptc…
# ℹ 88,118 more rowsLet’s try to load it
# A tibble: 88,128 × 5
row col expected actual file
<int> <chr> <chr> <chr> <chr>
1 2755 Year 1/0/T/F/TRUE/FALSE 2020 '/Users/kjhealy/Documents/courses/mptc…
2 2756 Year 1/0/T/F/TRUE/FALSE 2020 '/Users/kjhealy/Documents/courses/mptc…
3 2757 Year 1/0/T/F/TRUE/FALSE 2020 '/Users/kjhealy/Documents/courses/mptc…
4 2758 Year 1/0/T/F/TRUE/FALSE 2020 '/Users/kjhealy/Documents/courses/mptc…
5 2759 Year 1/0/T/F/TRUE/FALSE 2020 '/Users/kjhealy/Documents/courses/mptc…
6 2760 Year 1/0/T/F/TRUE/FALSE 2020 '/Users/kjhealy/Documents/courses/mptc…
7 2761 Year 1/0/T/F/TRUE/FALSE 2020 '/Users/kjhealy/Documents/courses/mptc…
8 2762 Year 1/0/T/F/TRUE/FALSE 2020 '/Users/kjhealy/Documents/courses/mptc…
9 2763 Year 1/0/T/F/TRUE/FALSE 2020 '/Users/kjhealy/Documents/courses/mptc…
10 2764 Year 1/0/T/F/TRUE/FALSE 2020 '/Users/kjhealy/Documents/courses/mptc…
# ℹ 88,118 more rows- Problems are stored as an attribute of the
nchsobject, so we can revisit them. - Parsing failures tend to cascade. Our data only has 56k rows but we got 88k failures.
Take a look with head()
# A tibble: 6 × 16
`Data As Of` `Start Date` `End Date` Group Year Month State Sex `Age Group`
<chr> <chr> <chr> <chr> <lgl> <lgl> <chr> <chr> <chr>
1 04/07/2021 01/01/2020 04/03/2021 By T… NA NA Unit… All … All Ages
2 04/07/2021 01/01/2020 04/03/2021 By T… NA NA Unit… All … Under 1 ye…
3 04/07/2021 01/01/2020 04/03/2021 By T… NA NA Unit… All … 0-17 years
4 04/07/2021 01/01/2020 04/03/2021 By T… NA NA Unit… All … 1-4 years
5 04/07/2021 01/01/2020 04/03/2021 By T… NA NA Unit… All … 5-14 years
6 04/07/2021 01/01/2020 04/03/2021 By T… NA NA Unit… All … 15-24 years
# ℹ 7 more variables: `COVID-19 Deaths` <dbl>, `Total Deaths` <dbl>,
# `Pneumonia Deaths` <dbl>, `Pneumonia and COVID-19 Deaths` <dbl>,
# `Influenza Deaths` <dbl>, `Pneumonia, Influenza, or COVID-19 Deaths` <dbl>,
# Footnote <chr>Take a look with tail()
# A tibble: 6 × 16
`Data As Of` `Start Date` `End Date` Group Year Month State Sex `Age Group`
<chr> <chr> <chr> <chr> <lgl> <lgl> <chr> <chr> <chr>
1 04/07/2021 04/01/2021 04/03/2021 By M… NA NA Puer… Fema… 45-54 years
2 04/07/2021 04/01/2021 04/03/2021 By M… NA NA Puer… Fema… 50-64 years
3 04/07/2021 04/01/2021 04/03/2021 By M… NA NA Puer… Fema… 55-64 years
4 04/07/2021 04/01/2021 04/03/2021 By M… NA NA Puer… Fema… 65-74 years
5 04/07/2021 04/01/2021 04/03/2021 By M… NA NA Puer… Fema… 75-84 years
6 04/07/2021 04/01/2021 04/03/2021 By M… NA NA Puer… Fema… 85 years a…
# ℹ 7 more variables: `COVID-19 Deaths` <dbl>, `Total Deaths` <dbl>,
# `Pneumonia Deaths` <dbl>, `Pneumonia and COVID-19 Deaths` <dbl>,
# `Influenza Deaths` <dbl>, `Pneumonia, Influenza, or COVID-19 Deaths` <dbl>,
# Footnote <chr>Take a look with slice_sample()
# A tibble: 10 × 16
`Data As Of` `Start Date` `End Date` Group Year Month State Sex
<chr> <chr> <chr> <chr> <lgl> <lgl> <chr> <chr>
1 04/07/2021 03/01/2020 03/31/2020 By Month NA NA Mississippi All …
2 04/07/2021 08/01/2020 08/31/2020 By Month NA NA New York All …
3 04/07/2021 05/01/2020 05/31/2020 By Month NA NA Idaho Fema…
4 04/07/2021 10/01/2020 10/31/2020 By Month NA NA South Caroli… Fema…
5 04/07/2021 01/01/2020 01/31/2020 By Month NA TRUE Vermont Fema…
6 04/07/2021 01/01/2021 01/31/2021 By Month NA TRUE New York City Male
7 04/07/2021 08/01/2020 08/31/2020 By Month NA NA Ohio Male
8 04/07/2021 10/01/2020 10/31/2020 By Month NA NA South Dakota Fema…
9 04/07/2021 02/01/2021 02/28/2021 By Month NA NA Kentucky Fema…
10 04/07/2021 10/01/2020 10/31/2020 By Month NA NA Alabama Fema…
# ℹ 8 more variables: `Age Group` <chr>, `COVID-19 Deaths` <dbl>,
# `Total Deaths` <dbl>, `Pneumonia Deaths` <dbl>,
# `Pneumonia and COVID-19 Deaths` <dbl>, `Influenza Deaths` <dbl>,
# `Pneumonia, Influenza, or COVID-19 Deaths` <dbl>, Footnote <chr>Aside: one that happened earlier …
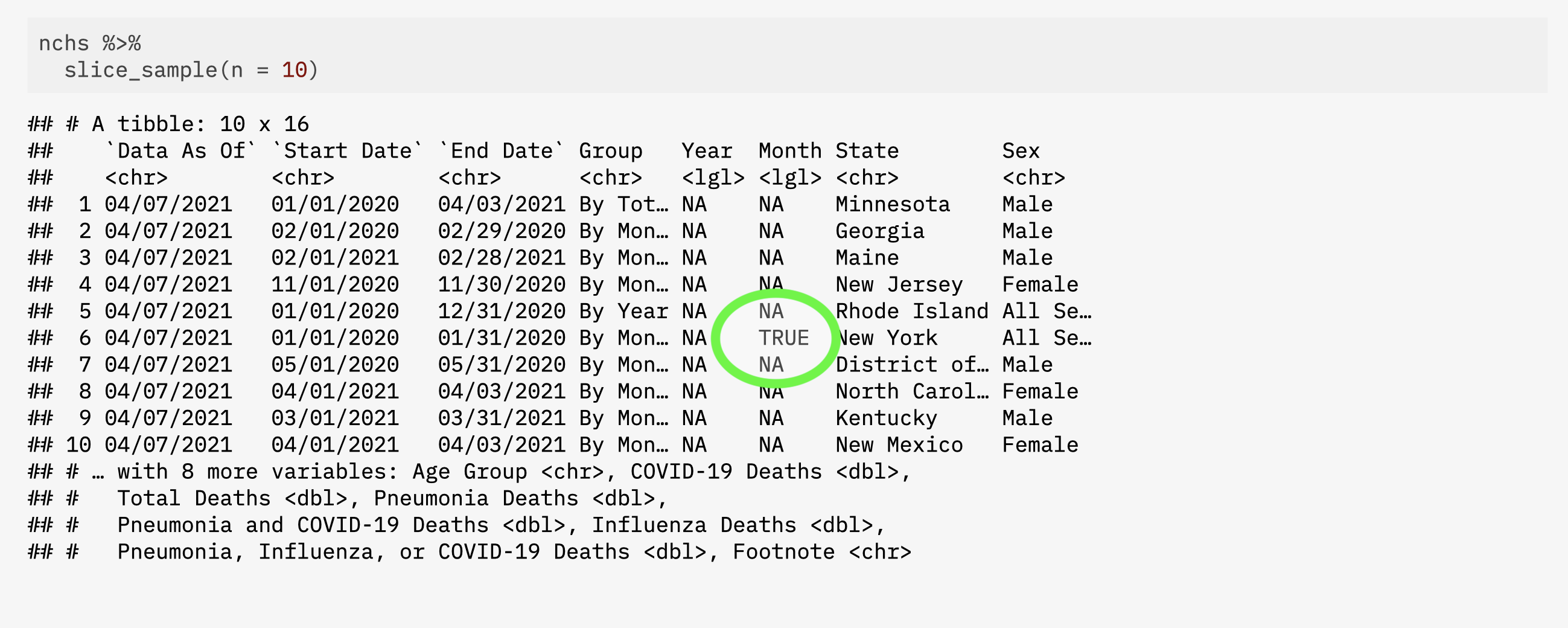
Take a look with slice()
- Let’s look at the rows
read_csv()complained about.
# A tibble: 11 × 16
`Data As Of` `Start Date` `End Date` Group Year Month State Sex
<chr> <chr> <chr> <chr> <lgl> <lgl> <chr> <chr>
1 04/07/2021 01/01/2020 04/03/2021 By Total NA NA Puerto Rico Fema…
2 04/07/2021 01/01/2020 04/03/2021 By Total NA NA Puerto Rico Fema…
3 04/07/2021 01/01/2020 04/03/2021 By Total NA NA Puerto Rico Fema…
4 04/07/2021 01/01/2020 04/03/2021 By Total NA NA Puerto Rico Fema…
5 04/07/2021 01/01/2020 04/03/2021 By Total NA NA Puerto Rico Fema…
6 04/07/2021 01/01/2020 12/31/2020 By Year NA NA United States All …
7 04/07/2021 01/01/2020 12/31/2020 By Year NA NA United States All …
8 04/07/2021 01/01/2020 12/31/2020 By Year NA NA United States All …
9 04/07/2021 01/01/2020 12/31/2020 By Year NA NA United States All …
10 04/07/2021 01/01/2020 12/31/2020 By Year NA NA United States All …
11 04/07/2021 01/01/2020 12/31/2020 By Year NA NA United States All …
# ℹ 8 more variables: `Age Group` <chr>, `COVID-19 Deaths` <dbl>,
# `Total Deaths` <dbl>, `Pneumonia Deaths` <dbl>,
# `Pneumonia and COVID-19 Deaths` <dbl>, `Influenza Deaths` <dbl>,
# `Pneumonia, Influenza, or COVID-19 Deaths` <dbl>, Footnote <chr>Take a look with slice()
# A tibble: 11 × 3
Year Month State
<lgl> <lgl> <chr>
1 NA NA Puerto Rico
2 NA NA Puerto Rico
3 NA NA Puerto Rico
4 NA NA Puerto Rico
5 NA NA Puerto Rico
6 NA NA United States
7 NA NA United States
8 NA NA United States
9 NA NA United States
10 NA NA United States
11 NA NA United States- Hm, something to do with the transition to national numbers maybe?
Take a look with select() & filter()
# A tibble: 969 × 3
Year Month State
<lgl> <lgl> <chr>
1 NA NA New York
2 NA NA New York
3 NA NA New York
4 NA NA New York
5 NA NA New York
6 NA NA New York
7 NA NA New York
8 NA NA New York
9 NA NA New York
10 NA NA New York
# ℹ 959 more rowsTake a look with is.na()
# A tibble: 0 × 3
# ℹ 3 variables: Year <lgl>, Month <lgl>, State <chr>- It really has been read in as a completely empty column.
- That doesn’t seem like it can be right.
Take a look with distinct()
- Again, it’s been read in as a completely empty column.
Take a look with read_lines()
- Time to reach for a different kitchen knife.
[1] "Data As Of,Start Date,End Date,Group,Year,Month,State,Sex,Age Group,COVID-19 Deaths,Total Deaths,Pneumonia Deaths,Pneumonia and COVID-19 Deaths,Influenza Deaths,\"Pneumonia, Influenza, or COVID-19 Deaths\",Footnote"
[2] "04/07/2021,01/01/2020,04/03/2021,By Total,,,United States,All Sexes,All Ages,539723,4161167,466437,263147,9037,750804,"
[3] "04/07/2021,01/01/2020,04/03/2021,By Total,,,United States,All Sexes,Under 1 year,59,22626,246,10,21,316,"
[4] "04/07/2021,01/01/2020,04/03/2021,By Total,,,United States,All Sexes,0-17 years,251,39620,667,46,179,1051,"
[5] "04/07/2021,01/01/2020,04/03/2021,By Total,,,United States,All Sexes,1-4 years,31,4069,137,5,61,224,"
[6] "04/07/2021,01/01/2020,04/03/2021,By Total,,,United States,All Sexes,5-14 years,89,6578,195,19,76,341,"
[7] "04/07/2021,01/01/2020,04/03/2021,By Total,,,United States,All Sexes,15-24 years,804,42596,930,317,81,1493,"
[8] "04/07/2021,01/01/2020,04/03/2021,By Total,,,United States,All Sexes,18-29 years,1996,75339,2184,884,150,3434,"
[9] "04/07/2021,01/01/2020,04/03/2021,By Total,,,United States,All Sexes,25-34 years,3543,88196,3493,1617,237,5638,"
[10] "04/07/2021,01/01/2020,04/03/2021,By Total,,,United States,All Sexes,30-39 years,5792,107348,5276,2658,318,8706," We can get the whole thing this way
- This imports the data as a long, long character vector, with each element being a line.
Now we’re just looking at lines in a file
# This is not a tibble; it's a vector. We have to index it the basic way, like letters[1:5]
raw_file[2753:2758] [1] "04/07/2021,01/01/2020,04/03/2021,By Total,,,Puerto Rico,Female,65-74 years,203,2650,410,151,,466,One or more data cells have counts between 1-9 and have been suppressed in accordance with NCHS confidentiality standards."
[2] "04/07/2021,01/01/2020,04/03/2021,By Total,,,Puerto Rico,Female,75-84 years,234,4274,656,154,16,751,"
[3] "04/07/2021,01/01/2020,04/03/2021,By Total,,,Puerto Rico,Female,85 years and over,222,6164,795,136,29,909,"
[4] "04/07/2021,01/01/2020,12/31/2020,By Year,2020,,United States,All Sexes,All Ages,380949,3372967,349667,178222,8779,560025,"
[5] "04/07/2021,01/01/2020,12/31/2020,By Year,2020,,United States,All Sexes,Under 1 year,48,19356,224,9,21,284,"
[6] "04/07/2021,01/01/2020,12/31/2020,By Year,2020,,United States,All Sexes,0-17 years,189,33808,598,35,178,930," There you are, you bastard.
In this case, this is due to the kind of data this is, mixing multiple reporting levels and totals. That is, it’s not a mistake in the data, but rather in the parsing.
OK, let’s go back to the colspec!
── Column specification ────────────────────────────────────────────────────────
cols(
`Data As Of` = col_character(),
`Start Date` = col_character(),
`End Date` = col_character(),
Group = col_character(),
Year = col_logical(),
Month = col_logical(),
State = col_character(),
Sex = col_character(),
`Age Group` = col_character(),
`COVID-19 Deaths` = col_double(),
`Total Deaths` = col_double(),
`Pneumonia Deaths` = col_double(),
`Pneumonia and COVID-19 Deaths` = col_double(),
`Influenza Deaths` = col_double(),
`Pneumonia, Influenza, or COVID-19 Deaths` = col_double(),
Footnote = col_character()
)- We can just copy it from the console output! It’s valid code.
We use it with col_types
nchs <- with_edition(1, read_csv(here("files", "examples", "SAS_on_2021-04-13.csv"),
col_types = cols(
`Data As Of` = col_character(),
`Start Date` = col_character(),
`End Date` = col_character(),
Group = col_character(),
Year = col_logical(),
Month = col_logical(),
State = col_character(),
Sex = col_character(),
`Age Group` = col_character(),
`COVID-19 Deaths` = col_double(),
`Total Deaths` = col_double(),
`Pneumonia Deaths` = col_double(),
`Pneumonia and COVID-19 Deaths` = col_double(),
`Influenza Deaths` = col_double(),
`Pneumonia, Influenza, or COVID-19 Deaths` = col_double(),
Footnote = col_character()
)))- But we know we need to make some adjustments.
Fixes
# Date format
us_style <- "%m/%d/%Y"
nchs <- with_edition(1, read_csv(
here("files", "examples", "SAS_on_2021-04-13.csv"),
col_types = cols(
`Data As Of` = col_date(format = us_style),
`Start Date` = col_date(format = us_style),
`End Date` = col_date(format = us_style),
Group = col_character(),
Year = col_character(),
Month = col_character(),
State = col_character(),
Sex = col_character(),
`Age Group` = col_character(),
`COVID-19 Deaths` = col_integer(),
`Total Deaths` = col_integer(),
`Pneumonia Deaths` = col_integer(),
`Pneumonia and COVID-19 Deaths` = col_integer(),
`Influenza Deaths` = col_integer(),
`Pneumonia, Influenza, or COVID-19 Deaths` = col_integer(),
Footnote = col_character()
)) |>
janitor::clean_names() |>
select(-footnote) |>
mutate(age_group = str_to_sentence(age_group)) |>
filter(!str_detect(state, "Total"))
)Now let’s look again
[1] 52326 15# A tibble: 49,572 × 3
year month state
<chr> <chr> <chr>
1 2020 <NA> United States
2 2020 <NA> United States
3 2020 <NA> United States
4 2020 <NA> United States
5 2020 <NA> United States
6 2020 <NA> United States
7 2020 <NA> United States
8 2020 <NA> United States
9 2020 <NA> United States
10 2020 <NA> United States
# ℹ 49,562 more rowsNow let’s look again
Lessons learned
- I said at the start that it was no fun, but also weirdly satisfying.
- When
read_csv()warns you of a parsing failure, don’t ignore it. read_lines()lets you get the file in a nearly unprocessed form.- The
colspecoutput is your friend.
If we wanted to …
If we wanted to …
# A tibble: 52,326 × 15
data_as_of start_date end_date group year month state sex age_group
<date> <date> <date> <chr> <chr> <chr> <chr> <chr> <chr>
1 2021-04-07 2020-01-01 2021-04-03 By Total <NA> <NA> United… All … All ages
2 2021-04-07 2020-01-01 2021-04-03 By Total <NA> <NA> United… All … Under 1 …
3 2021-04-07 2020-01-01 2021-04-03 By Total <NA> <NA> United… All … 0-17 yea…
4 2021-04-07 2020-01-01 2021-04-03 By Total <NA> <NA> United… All … 1-4 years
5 2021-04-07 2020-01-01 2021-04-03 By Total <NA> <NA> United… All … 5-14 yea…
6 2021-04-07 2020-01-01 2021-04-03 By Total <NA> <NA> United… All … 15-24 ye…
7 2021-04-07 2020-01-01 2021-04-03 By Total <NA> <NA> United… All … 18-29 ye…
8 2021-04-07 2020-01-01 2021-04-03 By Total <NA> <NA> United… All … 25-34 ye…
9 2021-04-07 2020-01-01 2021-04-03 By Total <NA> <NA> United… All … 30-39 ye…
10 2021-04-07 2020-01-01 2021-04-03 By Total <NA> <NA> United… All … 35-44 ye…
# ℹ 52,316 more rows
# ℹ 6 more variables: covid_19_deaths <int>, total_deaths <int>,
# pneumonia_deaths <int>, pneumonia_and_covid_19_deaths <int>,
# influenza_deaths <int>, pneumonia_influenza_or_covid_19_deaths <int>If we wanted to …
# A tibble: 52,326 × 10
group state sex age_group covid_19_deaths total_deaths pneumonia_deaths
<chr> <chr> <chr> <chr> <int> <int> <int>
1 By Total Unite… All … All ages 539723 4161167 466437
2 By Total Unite… All … Under 1 … 59 22626 246
3 By Total Unite… All … 0-17 yea… 251 39620 667
4 By Total Unite… All … 1-4 years 31 4069 137
5 By Total Unite… All … 5-14 yea… 89 6578 195
6 By Total Unite… All … 15-24 ye… 804 42596 930
7 By Total Unite… All … 18-29 ye… 1996 75339 2184
8 By Total Unite… All … 25-34 ye… 3543 88196 3493
9 By Total Unite… All … 30-39 ye… 5792 107348 5276
10 By Total Unite… All … 35-44 ye… 9259 126848 8203
# ℹ 52,316 more rows
# ℹ 3 more variables: pneumonia_and_covid_19_deaths <int>,
# influenza_deaths <int>, pneumonia_influenza_or_covid_19_deaths <int>If we wanted to …
# A tibble: 313,956 × 6
group state sex age_group outcome n
<chr> <chr> <chr> <chr> <chr> <int>
1 By Total United States All Sexes All ages covid_19_deaths 5.40e5
2 By Total United States All Sexes All ages total_deaths 4.16e6
3 By Total United States All Sexes All ages pneumonia_deaths 4.66e5
4 By Total United States All Sexes All ages pneumonia_and_covid_19_… 2.63e5
5 By Total United States All Sexes All ages influenza_deaths 9.04e3
6 By Total United States All Sexes All ages pneumonia_influenza_or_… 7.51e5
7 By Total United States All Sexes Under 1 year covid_19_deaths 5.9 e1
8 By Total United States All Sexes Under 1 year total_deaths 2.26e4
9 By Total United States All Sexes Under 1 year pneumonia_deaths 2.46e2
10 By Total United States All Sexes Under 1 year pneumonia_and_covid_19_… 1 e1
# ℹ 313,946 more rowsIf we wanted to …
library(stringr) # it's back!
nchs |>
select(!(c(data_as_of:end_date, year, month))) |>
pivot_longer(covid_19_deaths:pneumonia_influenza_or_covid_19_deaths,
names_to = "outcome",
values_to = "n") |>
mutate(outcome = str_to_sentence(outcome),
outcome = str_replace_all(outcome, "_", " "),
outcome = str_replace(outcome, "(C|c)ovid 19", "COVID-19"))# A tibble: 313,956 × 6
group state sex age_group outcome n
<chr> <chr> <chr> <chr> <chr> <int>
1 By Total United States All Sexes All ages COVID-19 deaths 5.40e5
2 By Total United States All Sexes All ages Total deaths 4.16e6
3 By Total United States All Sexes All ages Pneumonia deaths 4.66e5
4 By Total United States All Sexes All ages Pneumonia and COVID-19 … 2.63e5
5 By Total United States All Sexes All ages Influenza deaths 9.04e3
6 By Total United States All Sexes All ages Pneumonia influenza or … 7.51e5
7 By Total United States All Sexes Under 1 year COVID-19 deaths 5.9 e1
8 By Total United States All Sexes Under 1 year Total deaths 2.26e4
9 By Total United States All Sexes Under 1 year Pneumonia deaths 2.46e2
10 By Total United States All Sexes Under 1 year Pneumonia and COVID-19 … 1 e1
# ℹ 313,946 more rowsIf we wanted to …
- Put this in an object called
nchs_fmt
… we could make a table or graph
# A tibble: 313,956 × 4
state age_group outcome n
<chr> <chr> <chr> <int>
1 United States All ages COVID-19 deaths 539723
2 United States All ages Total deaths 4161167
3 United States All ages Pneumonia deaths 466437
4 United States All ages Pneumonia and COVID-19 deaths 263147
5 United States All ages Influenza deaths 9037
6 United States All ages Pneumonia influenza or COVID-19 deaths 750804
7 United States Under 1 year COVID-19 deaths 59
8 United States Under 1 year Total deaths 22626
9 United States Under 1 year Pneumonia deaths 246
10 United States Under 1 year Pneumonia and COVID-19 deaths 10
# ℹ 313,946 more rowsCleaned up (but not tidy)
Cleaned up (but not tidy)
# A tibble: 17 × 1
age_group
<chr>
1 All ages
2 Under 1 year
3 0-17 years
4 1-4 years
5 5-14 years
6 15-24 years
7 18-29 years
8 25-34 years
9 30-39 years
10 35-44 years
11 40-49 years
12 45-54 years
13 50-64 years
14 55-64 years
15 65-74 years
16 75-84 years
17 85 years and overMake our plot
p_out <- nchs_fmt |>
filter(group %in% "By Total",
sex %in% "All Sexes",
state %in% "United States",
age_group %in% c("0-17 years",
"18-29 years",
"30-39 years",
"40-49 years",
"50-64 years",
"65-74 years",
"85 years and over"),
outcome %in% "COVID-19 deaths") |>
mutate(age_group = str_replace(age_group, "years", "yrs"),
age_group = str_replace(age_group, " and over", ""),
age_group = str_replace(age_group, "85", "85+")) |>
ggplot(mapping = aes(x = n, y = age_group)) +
geom_col() + scale_x_continuous(labels = scales::comma) +
labs(x = "Deaths", y = NULL, title = "U.S. COVID-19 mortality totals by age group")Result

Every dataset is different
Dropping missings
Dropping missing values
Dropping missing values
- Drops all rows with any missing cases.
Dropping missing values
- What if we only want to drop all rows with all missing cases?
Revisit: Using across() and where()
Do more than one thing
A few weeks ago we saw this:
gss_sm |>
group_by(race, sex, degree) |>
summarize(n = n(),
mean_age = mean(age, na.rm = TRUE),
mean_kids = mean(childs, na.rm = TRUE))# A tibble: 34 × 6
# Groups: race, sex [6]
race sex degree n mean_age mean_kids
<fct> <fct> <fct> <int> <dbl> <dbl>
1 White Male Lt High School 96 52.9 2.45
2 White Male High School 470 48.8 1.61
3 White Male Junior College 65 47.1 1.54
4 White Male Bachelor 208 48.6 1.35
5 White Male Graduate 112 56.0 1.71
6 White Female Lt High School 101 55.4 2.81
7 White Female High School 587 51.9 1.98
8 White Female Junior College 101 48.2 1.91
9 White Female Bachelor 218 49.2 1.44
10 White Female Graduate 138 53.6 1.38
# ℹ 24 more rowsDo more than one thing
Similarly for organdata we might want to do:
organdata |>
group_by(consent_law, country) |>
summarize(donors_mean = mean(donors, na.rm = TRUE),
donors_sd = sd(donors, na.rm = TRUE),
gdp_mean = mean(gdp, na.rm = TRUE),
health_mean = mean(health, na.rm = TRUE),
roads_mean = mean(roads, na.rm = TRUE))# A tibble: 17 × 7
# Groups: consent_law [2]
consent_law country donors_mean donors_sd gdp_mean health_mean roads_mean
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Informed Australia 10.6 1.14 22179. 1958. 105.
2 Informed Canada 14.0 0.751 23711. 2272. 109.
3 Informed Denmark 13.1 1.47 23722. 2054. 102.
4 Informed Germany 13.0 0.611 22163. 2349. 113.
5 Informed Ireland 19.8 2.48 20824. 1480. 118.
6 Informed Netherlands 13.7 1.55 23013. 1993. 76.1
7 Informed United Kin… 13.5 0.775 21359. 1561. 67.9
8 Informed United Sta… 20.0 1.33 29212. 3988. 155.
9 Presumed Austria 23.5 2.42 23876. 1875. 150.
10 Presumed Belgium 21.9 1.94 22500. 1958. 155.
11 Presumed Finland 18.4 1.53 21019. 1615. 93.6
12 Presumed France 16.8 1.60 22603. 2160. 156.
13 Presumed Italy 11.1 4.28 21554. 1757 122.
14 Presumed Norway 15.4 1.11 26448. 2217. 70.0
15 Presumed Spain 28.1 4.96 16933 1289. 161.
16 Presumed Sweden 13.1 1.75 22415. 1951. 72.3
17 Presumed Switzerland 14.2 1.71 27233 2776. 96.4This works, but it’s really tedious. Also error-prone.
Use across()
Instead, use across() to apply a function to more than one column.
my_vars <- c("gdp", "donors", "roads")
## nested parens again, but it's worth it
organdata |>
group_by(consent_law, country) |>
summarize(across(all_of(my_vars),
list(avg = \(x) mean(x, na.rm = TRUE))
)
) # A tibble: 17 × 5
# Groups: consent_law [2]
consent_law country gdp_avg donors_avg roads_avg
<chr> <chr> <dbl> <dbl> <dbl>
1 Informed Australia 22179. 10.6 105.
2 Informed Canada 23711. 14.0 109.
3 Informed Denmark 23722. 13.1 102.
4 Informed Germany 22163. 13.0 113.
5 Informed Ireland 20824. 19.8 118.
6 Informed Netherlands 23013. 13.7 76.1
7 Informed United Kingdom 21359. 13.5 67.9
8 Informed United States 29212. 20.0 155.
9 Presumed Austria 23876. 23.5 150.
10 Presumed Belgium 22500. 21.9 155.
11 Presumed Finland 21019. 18.4 93.6
12 Presumed France 22603. 16.8 156.
13 Presumed Italy 21554. 11.1 122.
14 Presumed Norway 26448. 15.4 70.0
15 Presumed Spain 16933 28.1 161.
16 Presumed Sweden 22415. 13.1 72.3
17 Presumed Switzerland 27233 14.2 96.4Let’s look at that again
Let’s look at that again
# A tibble: 238 × 21
country year donors pop pop_dens gdp gdp_lag health health_lag
<chr> <date> <dbl> <int> <dbl> <int> <int> <dbl> <dbl>
1 Australia NA NA 17065 0.220 16774 16591 1300 1224
2 Australia 1991-01-01 12.1 17284 0.223 17171 16774 1379 1300
3 Australia 1992-01-01 12.4 17495 0.226 17914 17171 1455 1379
4 Australia 1993-01-01 12.5 17667 0.228 18883 17914 1540 1455
5 Australia 1994-01-01 10.2 17855 0.231 19849 18883 1626 1540
6 Australia 1995-01-01 10.2 18072 0.233 21079 19849 1737 1626
7 Australia 1996-01-01 10.6 18311 0.237 21923 21079 1846 1737
8 Australia 1997-01-01 10.3 18518 0.239 22961 21923 1948 1846
9 Australia 1998-01-01 10.5 18711 0.242 24148 22961 2077 1948
10 Australia 1999-01-01 8.67 18926 0.244 25445 24148 2231 2077
# ℹ 228 more rows
# ℹ 12 more variables: pubhealth <dbl>, roads <dbl>, cerebvas <int>,
# assault <int>, external <int>, txp_pop <dbl>, world <chr>, opt <chr>,
# consent_law <chr>, consent_practice <chr>, consistent <chr>, ccode <chr>Let’s look at that again
# A tibble: 238 × 21
# Groups: consent_law, country [17]
country year donors pop pop_dens gdp gdp_lag health health_lag
<chr> <date> <dbl> <int> <dbl> <int> <int> <dbl> <dbl>
1 Australia NA NA 17065 0.220 16774 16591 1300 1224
2 Australia 1991-01-01 12.1 17284 0.223 17171 16774 1379 1300
3 Australia 1992-01-01 12.4 17495 0.226 17914 17171 1455 1379
4 Australia 1993-01-01 12.5 17667 0.228 18883 17914 1540 1455
5 Australia 1994-01-01 10.2 17855 0.231 19849 18883 1626 1540
6 Australia 1995-01-01 10.2 18072 0.233 21079 19849 1737 1626
7 Australia 1996-01-01 10.6 18311 0.237 21923 21079 1846 1737
8 Australia 1997-01-01 10.3 18518 0.239 22961 21923 1948 1846
9 Australia 1998-01-01 10.5 18711 0.242 24148 22961 2077 1948
10 Australia 1999-01-01 8.67 18926 0.244 25445 24148 2231 2077
# ℹ 228 more rows
# ℹ 12 more variables: pubhealth <dbl>, roads <dbl>, cerebvas <int>,
# assault <int>, external <int>, txp_pop <dbl>, world <chr>, opt <chr>,
# consent_law <chr>, consent_practice <chr>, consistent <chr>, ccode <chr>Let’s look at that again
# A tibble: 17 × 5
# Groups: consent_law [2]
consent_law country gdp_avg donors_avg roads_avg
<chr> <chr> <dbl> <dbl> <dbl>
1 Informed Australia 22179. 10.6 105.
2 Informed Canada 23711. 14.0 109.
3 Informed Denmark 23722. 13.1 102.
4 Informed Germany 22163. 13.0 113.
5 Informed Ireland 20824. 19.8 118.
6 Informed Netherlands 23013. 13.7 76.1
7 Informed United Kingdom 21359. 13.5 67.9
8 Informed United States 29212. 20.0 155.
9 Presumed Austria 23876. 23.5 150.
10 Presumed Belgium 22500. 21.9 155.
11 Presumed Finland 21019. 18.4 93.6
12 Presumed France 22603. 16.8 156.
13 Presumed Italy 21554. 11.1 122.
14 Presumed Norway 26448. 15.4 70.0
15 Presumed Spain 16933 28.1 161.
16 Presumed Sweden 22415. 13.1 72.3
17 Presumed Switzerland 27233 14.2 96.4Let’s look at that again
my_varsare selected byacross()- We use
all_of()orany_of()to be explicit list()of the formresult = functiongives the new columns that will be calculated.- The thing inside the list with the “waving person”,
\(x), is an anonymous function
We can calculate more than one thing
my_vars <- c("gdp", "donors", "roads")
organdata |>
group_by(consent_law, country) |>
summarize(across(all_of(my_vars),
list(avg = \(x) mean(x, na.rm = TRUE),
sdev = \(x) sd(x, na.rm = TRUE),
md = \(x) median(x, na.rm = TRUE))
)
)# A tibble: 17 × 11
# Groups: consent_law [2]
consent_law country gdp_avg gdp_sdev gdp_md donors_avg donors_sdev donors_md
<chr> <chr> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
1 Informed Austral… 22179. 3959. 21923 10.6 1.14 10.4
2 Informed Canada 23711. 3966. 22764 14.0 0.751 14.0
3 Informed Denmark 23722. 3896. 23548 13.1 1.47 12.9
4 Informed Germany 22163. 2501. 22164 13.0 0.611 13
5 Informed Ireland 20824. 6670. 19245 19.8 2.48 19.2
6 Informed Netherl… 23013. 3770. 22541 13.7 1.55 13.8
7 Informed United … 21359. 3929. 20839 13.5 0.775 13.5
8 Informed United … 29212. 4571. 28772 20.0 1.33 20.1
9 Presumed Austria 23876. 3343. 23798 23.5 2.42 23.8
10 Presumed Belgium 22500. 3171. 22152 21.9 1.94 21.4
11 Presumed Finland 21019. 3668. 19842 18.4 1.53 19.4
12 Presumed France 22603. 3260. 21990 16.8 1.60 16.6
13 Presumed Italy 21554. 2781. 21396 11.1 4.28 11.3
14 Presumed Norway 26448. 6492. 26218 15.4 1.11 15.4
15 Presumed Spain 16933 2888. 16416 28.1 4.96 28
16 Presumed Sweden 22415. 3213. 22029 13.1 1.75 12.7
17 Presumed Switzer… 27233 2153. 26304 14.2 1.71 14.4
# ℹ 3 more variables: roads_avg <dbl>, roads_sdev <dbl>, roads_md <dbl>It’s OK to use the function names
my_vars <- c("gdp", "donors", "roads")
organdata |>
group_by(consent_law, country) |>
summarize(across(all_of(my_vars),
list(mean = \(x) mean(x, na.rm = TRUE),
sd = \(x) sd(x, na.rm = TRUE),
median = \(x) median(x, na.rm = TRUE))
)
)# A tibble: 17 × 11
# Groups: consent_law [2]
consent_law country gdp_mean gdp_sd gdp_median donors_mean donors_sd
<chr> <chr> <dbl> <dbl> <int> <dbl> <dbl>
1 Informed Australia 22179. 3959. 21923 10.6 1.14
2 Informed Canada 23711. 3966. 22764 14.0 0.751
3 Informed Denmark 23722. 3896. 23548 13.1 1.47
4 Informed Germany 22163. 2501. 22164 13.0 0.611
5 Informed Ireland 20824. 6670. 19245 19.8 2.48
6 Informed Netherlands 23013. 3770. 22541 13.7 1.55
7 Informed United Kingdom 21359. 3929. 20839 13.5 0.775
8 Informed United States 29212. 4571. 28772 20.0 1.33
9 Presumed Austria 23876. 3343. 23798 23.5 2.42
10 Presumed Belgium 22500. 3171. 22152 21.9 1.94
11 Presumed Finland 21019. 3668. 19842 18.4 1.53
12 Presumed France 22603. 3260. 21990 16.8 1.60
13 Presumed Italy 21554. 2781. 21396 11.1 4.28
14 Presumed Norway 26448. 6492. 26218 15.4 1.11
15 Presumed Spain 16933 2888. 16416 28.1 4.96
16 Presumed Sweden 22415. 3213. 22029 13.1 1.75
17 Presumed Switzerland 27233 2153. 26304 14.2 1.71
# ℹ 4 more variables: donors_median <dbl>, roads_mean <dbl>, roads_sd <dbl>,
# roads_median <dbl>Selection with across(where())
organdata |>
group_by(consent_law, country) |>
summarize(across(where(is.numeric),
list(mean = \(x) mean(x, na.rm = TRUE),
sd = \(x) sd(x, na.rm = TRUE),
median = \(x) median(x, na.rm = TRUE))
)
) |>
print(n = 3) # just to save slide space# A tibble: 17 × 41
# Groups: consent_law [2]
consent_law country donors_mean donors_sd donors_median pop_mean pop_sd
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Informed Australia 10.6 1.14 10.4 18318. 831.
2 Informed Canada 14.0 0.751 14.0 29608. 1193.
3 Informed Denmark 13.1 1.47 12.9 5257. 80.6
# ℹ 14 more rows
# ℹ 34 more variables: pop_median <int>, pop_dens_mean <dbl>,
# pop_dens_sd <dbl>, pop_dens_median <dbl>, gdp_mean <dbl>, gdp_sd <dbl>,
# gdp_median <int>, gdp_lag_mean <dbl>, gdp_lag_sd <dbl>,
# gdp_lag_median <dbl>, health_mean <dbl>, health_sd <dbl>,
# health_median <dbl>, health_lag_mean <dbl>, health_lag_sd <dbl>,
# health_lag_median <dbl>, pubhealth_mean <dbl>, pubhealth_sd <dbl>, …Name new columns with .names
organdata |>
group_by(consent_law, country) |>
summarize(across(where(is.numeric),
list(mean = \(x) mean(x, na.rm = TRUE),
sd = \(x) sd(x, na.rm = TRUE),
median = \(x) median(x, na.rm = TRUE)),
.names = "{fn}_{col}"
)
) |>
print(n = 3) # A tibble: 17 × 41
# Groups: consent_law [2]
consent_law country mean_donors sd_donors median_donors mean_pop sd_pop
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Informed Australia 10.6 1.14 10.4 18318. 831.
2 Informed Canada 14.0 0.751 14.0 29608. 1193.
3 Informed Denmark 13.1 1.47 12.9 5257. 80.6
# ℹ 14 more rows
# ℹ 34 more variables: median_pop <int>, mean_pop_dens <dbl>,
# sd_pop_dens <dbl>, median_pop_dens <dbl>, mean_gdp <dbl>, sd_gdp <dbl>,
# median_gdp <int>, mean_gdp_lag <dbl>, sd_gdp_lag <dbl>,
# median_gdp_lag <dbl>, mean_health <dbl>, sd_health <dbl>,
# median_health <dbl>, mean_health_lag <dbl>, sd_health_lag <dbl>,
# median_health_lag <dbl>, mean_pubhealth <dbl>, sd_pubhealth <dbl>, …Name new columns with .names
In tidyverse functions, arguments that begin with a “.” generally have it in order to avoid confusion with existing items, or are “pronouns” referring to e.g. “the name of the thing we’re currently talking about as we evaluate this function”.
This all works with mutate(), too
# A tibble: 238 × 7
country world opt consent_law consent_practice consistent ccode
<chr> <chr> <chr> <chr> <chr> <chr> <chr>
1 AUSTRALIA LIBERAL IN INFORMED INFORMED YES OZ
2 AUSTRALIA LIBERAL IN INFORMED INFORMED YES OZ
3 AUSTRALIA LIBERAL IN INFORMED INFORMED YES OZ
4 AUSTRALIA LIBERAL IN INFORMED INFORMED YES OZ
5 AUSTRALIA LIBERAL IN INFORMED INFORMED YES OZ
6 AUSTRALIA LIBERAL IN INFORMED INFORMED YES OZ
7 AUSTRALIA LIBERAL IN INFORMED INFORMED YES OZ
8 AUSTRALIA LIBERAL IN INFORMED INFORMED YES OZ
9 AUSTRALIA LIBERAL IN INFORMED INFORMED YES OZ
10 AUSTRALIA LIBERAL IN INFORMED INFORMED YES OZ
# ℹ 228 more rowsExample: cleaning a table
Cleaning a table
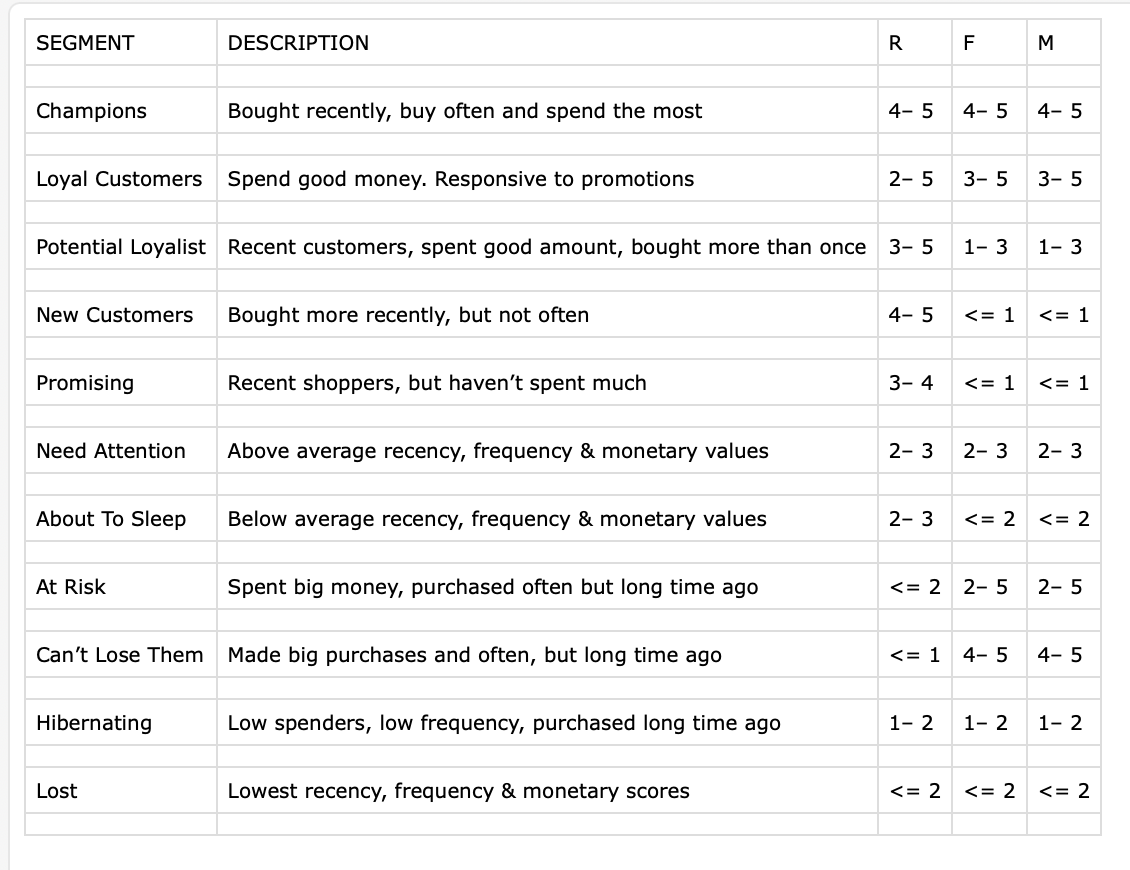
- With that in mind … Some marketing data
Cleaning a table
# A tibble: 23 × 5
SEGMENT DESCRIPTION R F M
<chr> <chr> <chr> <chr> <chr>
1 <NA> <NA> <NA> <NA> <NA>
2 Champions Bought recently, buy often and spend th… 4– 5 4– 5 4– 5
3 <NA> <NA> <NA> <NA> <NA>
4 Loyal Customers Spend good money. Responsive to promoti… 2– 5 3– 5 3– 5
5 <NA> <NA> <NA> <NA> <NA>
6 Potential Loyalist Recent customers, spent good amount, bo… 3– 5 1– 3 1– 3
7 <NA> <NA> <NA> <NA> <NA>
8 New Customers Bought more recently, but not often 4– 5 <= 1 <= 1
9 <NA> <NA> <NA> <NA> <NA>
10 Promising Recent shoppers, but haven’t spent much 3– 4 <= 1 <= 1
# ℹ 13 more rowsCleaning a table
# A tibble: 23 × 5
segment description r f m
<chr> <chr> <chr> <chr> <chr>
1 <NA> <NA> <NA> <NA> <NA>
2 Champions Bought recently, buy often and spend th… 4– 5 4– 5 4– 5
3 <NA> <NA> <NA> <NA> <NA>
4 Loyal Customers Spend good money. Responsive to promoti… 2– 5 3– 5 3– 5
5 <NA> <NA> <NA> <NA> <NA>
6 Potential Loyalist Recent customers, spent good amount, bo… 3– 5 1– 3 1– 3
7 <NA> <NA> <NA> <NA> <NA>
8 New Customers Bought more recently, but not often 4– 5 <= 1 <= 1
9 <NA> <NA> <NA> <NA> <NA>
10 Promising Recent shoppers, but haven’t spent much 3– 4 <= 1 <= 1
# ℹ 13 more rowsCleaning a table
# A tibble: 11 × 5
segment description r f m
<chr> <chr> <chr> <chr> <chr>
1 Champions Bought recently, buy often and spend th… 4– 5 4– 5 4– 5
2 Loyal Customers Spend good money. Responsive to promoti… 2– 5 3– 5 3– 5
3 Potential Loyalist Recent customers, spent good amount, bo… 3– 5 1– 3 1– 3
4 New Customers Bought more recently, but not often 4– 5 <= 1 <= 1
5 Promising Recent shoppers, but haven’t spent much 3– 4 <= 1 <= 1
6 Need Attention Above average recency, frequency & mone… 2– 3 2– 3 2– 3
7 About To Sleep Below average recency, frequency & mone… 2– 3 <= 2 <= 2
8 At Risk Spent big money, purchased often but lo… <= 2 2– 5 2– 5
9 Can’t Lose Them Made big purchases and often, but long … <= 1 4– 5 4– 5
10 Hibernating Low spenders, low frequency, purchased … 1– 2 1– 2 1– 2
11 Lost Lowest recency, frequency & monetary sc… <= 2 <= 2 <= 2 Cleaning a table
# A tibble: 33 × 4
segment description name value
<chr> <chr> <chr> <chr>
1 Champions Bought recently, buy often and spend the most r 4– 5
2 Champions Bought recently, buy often and spend the most f 4– 5
3 Champions Bought recently, buy often and spend the most m 4– 5
4 Loyal Customers Spend good money. Responsive to promotions r 2– 5
5 Loyal Customers Spend good money. Responsive to promotions f 3– 5
6 Loyal Customers Spend good money. Responsive to promotions m 3– 5
7 Potential Loyalist Recent customers, spent good amount, bought m… r 3– 5
8 Potential Loyalist Recent customers, spent good amount, bought m… f 1– 3
9 Potential Loyalist Recent customers, spent good amount, bought m… m 1– 3
10 New Customers Bought more recently, but not often r 4– 5
# ℹ 23 more rowsCleaning a table
# A tibble: 33 × 6
segment description name value lo hi
<chr> <chr> <chr> <chr> <int> <int>
1 Champions Bought recently, buy often and sp… r 4– 5 4 5
2 Champions Bought recently, buy often and sp… f 4– 5 4 5
3 Champions Bought recently, buy often and sp… m 4– 5 4 5
4 Loyal Customers Spend good money. Responsive to p… r 2– 5 2 5
5 Loyal Customers Spend good money. Responsive to p… f 3– 5 3 5
6 Loyal Customers Spend good money. Responsive to p… m 3– 5 3 5
7 Potential Loyalist Recent customers, spent good amou… r 3– 5 3 5
8 Potential Loyalist Recent customers, spent good amou… f 1– 3 1 3
9 Potential Loyalist Recent customers, spent good amou… m 1– 3 1 3
10 New Customers Bought more recently, but not oft… r 4– 5 4 5
# ℹ 23 more rowsCleaning a table
# A tibble: 33 × 5
segment description name lo hi
<chr> <chr> <chr> <int> <int>
1 Champions Bought recently, buy often and spend th… r 4 5
2 Champions Bought recently, buy often and spend th… f 4 5
3 Champions Bought recently, buy often and spend th… m 4 5
4 Loyal Customers Spend good money. Responsive to promoti… r 2 5
5 Loyal Customers Spend good money. Responsive to promoti… f 3 5
6 Loyal Customers Spend good money. Responsive to promoti… m 3 5
7 Potential Loyalist Recent customers, spent good amount, bo… r 3 5
8 Potential Loyalist Recent customers, spent good amount, bo… f 1 3
9 Potential Loyalist Recent customers, spent good amount, bo… m 1 3
10 New Customers Bought more recently, but not often r 4 5
# ℹ 23 more rowsCleaning a table
read_csv(here("files", "examples", "rfm_table.csv")) |>
janitor::clean_names() |>
janitor::remove_empty("rows") |>
pivot_longer(cols = r:m) |>
separate(col = value, into = c("lo", "hi"),
remove = FALSE, convert = TRUE,
fill = "left") |>
select(-value) |>
pivot_wider(names_from = name,
values_from = lo:hi)# A tibble: 11 × 8
segment description lo_r lo_f lo_m hi_r hi_f hi_m
<chr> <chr> <int> <int> <int> <int> <int> <int>
1 Champions Bought recently, buy … 4 4 4 5 5 5
2 Loyal Customers Spend good money. Res… 2 3 3 5 5 5
3 Potential Loyalist Recent customers, spe… 3 1 1 5 3 3
4 New Customers Bought more recently,… 4 NA NA 5 1 1
5 Promising Recent shoppers, but … 3 NA NA 4 1 1
6 Need Attention Above average recency… 2 2 2 3 3 3
7 About To Sleep Below average recency… 2 NA NA 3 2 2
8 At Risk Spent big money, purc… NA 2 2 2 5 5
9 Can’t Lose Them Made big purchases an… NA 4 4 1 5 5
10 Hibernating Low spenders, low fre… 1 1 1 2 2 2
11 Lost Lowest recency, frequ… NA NA NA 2 2 2Cleaning a table
read_csv(here("files", "examples", "rfm_table.csv")) |>
janitor::clean_names() |>
janitor::remove_empty("rows") |>
pivot_longer(cols = r:m) |>
separate(col = value, into = c("lo", "hi"),
remove = FALSE, convert = TRUE,
fill = "left") |>
select(-value) |>
pivot_wider(names_from = name,
values_from = lo:hi) |>
mutate(across(where(is.integer), replace_na, 0))# A tibble: 11 × 8
segment description lo_r lo_f lo_m hi_r hi_f hi_m
<chr> <chr> <int> <int> <int> <int> <int> <int>
1 Champions Bought recently, buy … 4 4 4 5 5 5
2 Loyal Customers Spend good money. Res… 2 3 3 5 5 5
3 Potential Loyalist Recent customers, spe… 3 1 1 5 3 3
4 New Customers Bought more recently,… 4 0 0 5 1 1
5 Promising Recent shoppers, but … 3 0 0 4 1 1
6 Need Attention Above average recency… 2 2 2 3 3 3
7 About To Sleep Below average recency… 2 0 0 3 2 2
8 At Risk Spent big money, purc… 0 2 2 2 5 5
9 Can’t Lose Them Made big purchases an… 0 4 4 1 5 5
10 Hibernating Low spenders, low fre… 1 1 1 2 2 2
11 Lost Lowest recency, frequ… 0 0 0 2 2 2Cleaning a table
read_csv(here("files", "examples", "rfm_table.csv")) |>
janitor::clean_names() |>
janitor::remove_empty("rows") |>
pivot_longer(cols = r:m) |>
separate(col = value, into = c("lo", "hi"),
remove = FALSE, convert = TRUE,
fill = "left") |>
select(-value) |>
pivot_wider(names_from = name,
values_from = lo:hi) |>
mutate(across(where(is.integer), replace_na, 0)) |>
select(segment,
lo_r, hi_r,
lo_f, hi_f,
lo_m, hi_m,
description)# A tibble: 11 × 8
segment lo_r hi_r lo_f hi_f lo_m hi_m description
<chr> <int> <int> <int> <int> <int> <int> <chr>
1 Champions 4 5 4 5 4 5 Bought recently, buy …
2 Loyal Customers 2 5 3 5 3 5 Spend good money. Res…
3 Potential Loyalist 3 5 1 3 1 3 Recent customers, spe…
4 New Customers 4 5 0 1 0 1 Bought more recently,…
5 Promising 3 4 0 1 0 1 Recent shoppers, but …
6 Need Attention 2 3 2 3 2 3 Above average recency…
7 About To Sleep 2 3 0 2 0 2 Below average recency…
8 At Risk 0 2 2 5 2 5 Spent big money, purc…
9 Can’t Lose Them 0 1 4 5 4 5 Made big purchases an…
10 Hibernating 1 2 1 2 1 2 Low spenders, low fre…
11 Lost 0 2 0 2 0 2 Lowest recency, frequ…A candidate for rowwise()?
# A tibble: 11 × 8
segment lo_r hi_r lo_f hi_f lo_m hi_m description
<chr> <int> <int> <int> <int> <int> <int> <chr>
1 Champions 4 5 4 5 4 5 Bought recently, buy …
2 Loyal Customers 2 5 3 5 3 5 Spend good money. Res…
3 Potential Loyalist 3 5 1 3 1 3 Recent customers, spe…
4 New Customers 4 5 0 1 0 1 Bought more recently,…
5 Promising 3 4 0 1 0 1 Recent shoppers, but …
6 Need Attention 2 3 2 3 2 3 Above average recency…
7 About To Sleep 2 3 0 2 0 2 Below average recency…
8 At Risk 0 2 2 5 2 5 Spent big money, purc…
9 Can’t Lose Them 0 1 4 5 4 5 Made big purchases an…
10 Hibernating 1 2 1 2 1 2 Low spenders, low fre…
11 Lost 0 2 0 2 0 2 Lowest recency, frequ…A candidate for rowwise()?
- This does what we expect:
rfm_table |>
mutate(sum_lo = lo_r + lo_f + lo_m,
sum_hi = hi_r + hi_f + hi_m) |>
select(segment, sum_lo, sum_hi, everything())# A tibble: 11 × 10
segment sum_lo sum_hi lo_r hi_r lo_f hi_f lo_m hi_m description
<chr> <int> <int> <int> <int> <int> <int> <int> <int> <chr>
1 Champions 12 15 4 5 4 5 4 5 Bought rec…
2 Loyal Customers 8 15 2 5 3 5 3 5 Spend good…
3 Potential Loya… 5 11 3 5 1 3 1 3 Recent cus…
4 New Customers 4 7 4 5 0 1 0 1 Bought mor…
5 Promising 3 6 3 4 0 1 0 1 Recent sho…
6 Need Attention 6 9 2 3 2 3 2 3 Above aver…
7 About To Sleep 2 7 2 3 0 2 0 2 Below aver…
8 At Risk 4 12 0 2 2 5 2 5 Spent big …
9 Can’t Lose Them 8 11 0 1 4 5 4 5 Made big p…
10 Hibernating 3 6 1 2 1 2 1 2 Low spende…
11 Lost 0 6 0 2 0 2 0 2 Lowest rec…This adds each column, elementwise.
A candidate for rowwise()?
- But this does not:
rfm_table |>
mutate(sum_lo = sum(lo_r, lo_f, lo_m),
sum_hi = sum(hi_r, hi_f, hi_m)) |>
select(segment, sum_lo, sum_hi, everything())# A tibble: 11 × 10
segment sum_lo sum_hi lo_r hi_r lo_f hi_f lo_m hi_m description
<chr> <int> <int> <int> <int> <int> <int> <int> <int> <chr>
1 Champions 55 105 4 5 4 5 4 5 Bought rec…
2 Loyal Customers 55 105 2 5 3 5 3 5 Spend good…
3 Potential Loya… 55 105 3 5 1 3 1 3 Recent cus…
4 New Customers 55 105 4 5 0 1 0 1 Bought mor…
5 Promising 55 105 3 4 0 1 0 1 Recent sho…
6 Need Attention 55 105 2 3 2 3 2 3 Above aver…
7 About To Sleep 55 105 2 3 0 2 0 2 Below aver…
8 At Risk 55 105 0 2 2 5 2 5 Spent big …
9 Can’t Lose Them 55 105 0 1 4 5 4 5 Made big p…
10 Hibernating 55 105 1 2 1 2 1 2 Low spende…
11 Lost 55 105 0 2 0 2 0 2 Lowest rec…- Sum is taking all the columns, adding them up (into a single number), and putting that result in each row.
A candidate for rowwise()?
- Similarly, this will not give the answer we probably expect:
rfm_table |>
mutate(mean_lo = mean(c(lo_r, lo_f, lo_m)),
mean_hi = mean(c(hi_r, hi_f, hi_m))) |>
select(segment, mean_lo, mean_hi, everything())# A tibble: 11 × 10
segment mean_lo mean_hi lo_r hi_r lo_f hi_f lo_m hi_m description
<chr> <dbl> <dbl> <int> <int> <int> <int> <int> <int> <chr>
1 Champions 1.67 3.18 4 5 4 5 4 5 Bought rec…
2 Loyal Custom… 1.67 3.18 2 5 3 5 3 5 Spend good…
3 Potential Lo… 1.67 3.18 3 5 1 3 1 3 Recent cus…
4 New Customers 1.67 3.18 4 5 0 1 0 1 Bought mor…
5 Promising 1.67 3.18 3 4 0 1 0 1 Recent sho…
6 Need Attenti… 1.67 3.18 2 3 2 3 2 3 Above aver…
7 About To Sle… 1.67 3.18 2 3 0 2 0 2 Below aver…
8 At Risk 1.67 3.18 0 2 2 5 2 5 Spent big …
9 Can’t Lose T… 1.67 3.18 0 1 4 5 4 5 Made big p…
10 Hibernating 1.67 3.18 1 2 1 2 1 2 Low spende…
11 Lost 1.67 3.18 0 2 0 2 0 2 Lowest rec…A candidate for rowwise()?
- But this will:
rfm_table |>
rowwise() |>
mutate(mean_lo = mean(c(lo_r, lo_f, lo_m)),
mean_hi = mean(c(hi_r, hi_f, hi_m))) |>
select(segment, mean_lo, mean_hi, everything())# A tibble: 11 × 10
# Rowwise:
segment mean_lo mean_hi lo_r hi_r lo_f hi_f lo_m hi_m description
<chr> <dbl> <dbl> <int> <int> <int> <int> <int> <int> <chr>
1 Champions 4 5 4 5 4 5 4 5 Bought rec…
2 Loyal Custom… 2.67 5 2 5 3 5 3 5 Spend good…
3 Potential Lo… 1.67 3.67 3 5 1 3 1 3 Recent cus…
4 New Customers 1.33 2.33 4 5 0 1 0 1 Bought mor…
5 Promising 1 2 3 4 0 1 0 1 Recent sho…
6 Need Attenti… 2 3 2 3 2 3 2 3 Above aver…
7 About To Sle… 0.667 2.33 2 3 0 2 0 2 Below aver…
8 At Risk 1.33 4 0 2 2 5 2 5 Spent big …
9 Can’t Lose T… 2.67 3.67 0 1 4 5 4 5 Made big p…
10 Hibernating 1 2 1 2 1 2 1 2 Low spende…
11 Lost 0 2 0 2 0 2 0 2 Lowest rec…Rowwise isn’t very efficient
- In general, you’ll want to see if some vectorized (“operating on columns, but elementwise”) function exists, as it’ll be faster.
- And most of the time, R and the tidyverse “wants” you to work in vectorized, columnar terms … hence your first move will often be to pivot the data into long format.
- So,
rowwise()is not likely to see a whole lot of further development.
You may want group_by() instead
rfm_table |>
group_by(segment) |>
mutate(mean_lo = mean(c(lo_r, lo_f, lo_m)),
mean_hi = mean(c(hi_r, hi_f, hi_m))) |>
select(segment, mean_lo, mean_hi, everything())# A tibble: 11 × 10
# Groups: segment [11]
segment mean_lo mean_hi lo_r hi_r lo_f hi_f lo_m hi_m description
<chr> <dbl> <dbl> <int> <int> <int> <int> <int> <int> <chr>
1 Champions 4 5 4 5 4 5 4 5 Bought rec…
2 Loyal Custom… 2.67 5 2 5 3 5 3 5 Spend good…
3 Potential Lo… 1.67 3.67 3 5 1 3 1 3 Recent cus…
4 New Customers 1.33 2.33 4 5 0 1 0 1 Bought mor…
5 Promising 1 2 3 4 0 1 0 1 Recent sho…
6 Need Attenti… 2 3 2 3 2 3 2 3 Above aver…
7 About To Sle… 0.667 2.33 2 3 0 2 0 2 Below aver…
8 At Risk 1.33 4 0 2 2 5 2 5 Spent big …
9 Can’t Lose T… 2.67 3.67 0 1 4 5 4 5 Made big p…
10 Hibernating 1 2 1 2 1 2 1 2 Low spende…
11 Lost 0 2 0 2 0 2 0 2 Lowest rec…You may want group_by() instead
rfm_table |>
group_by(segment) |>
mutate(sum_lo = sum(lo_r, lo_f, lo_m),
sum_hi = sum(hi_r, hi_f, hi_m)) |>
select(segment, sum_lo, sum_hi, everything())# A tibble: 11 × 10
# Groups: segment [11]
segment sum_lo sum_hi lo_r hi_r lo_f hi_f lo_m hi_m description
<chr> <int> <int> <int> <int> <int> <int> <int> <int> <chr>
1 Champions 12 15 4 5 4 5 4 5 Bought rec…
2 Loyal Customers 8 15 2 5 3 5 3 5 Spend good…
3 Potential Loya… 5 11 3 5 1 3 1 3 Recent cus…
4 New Customers 4 7 4 5 0 1 0 1 Bought mor…
5 Promising 3 6 3 4 0 1 0 1 Recent sho…
6 Need Attention 6 9 2 3 2 3 2 3 Above aver…
7 About To Sleep 2 7 2 3 0 2 0 2 Below aver…
8 At Risk 4 12 0 2 2 5 2 5 Spent big …
9 Can’t Lose Them 8 11 0 1 4 5 4 5 Made big p…
10 Hibernating 3 6 1 2 1 2 1 2 Low spende…
11 Lost 0 6 0 2 0 2 0 2 Lowest rec…Foreign formats
What about Stata?
Using haven
- Haven is the Tidyverse’s package for reading and managing files from Stata, SPSS, and SAS. You should prefer it to the older Base R package
foreign, which has similar functionality. - We’re going to import a General Social Survey dataset that’s in Stata’s
.dtaformat.
We’ll do some of the common recoding and reorganizing tasks that accompany this.
The GSS panel
- The data:
# A tibble: 14,610 × 2,757
firstyear firstid year id vpsu vstrat adults ballot dateintv famgen
<dbl> <dbl+lbl> <dbl> <dbl> <dbl+> <dbl+> <dbl+> <dbl+l> <dbl+lb> <dbl+l>
1 2006 9 2006 9 2 1957 1 3 [BAL… 709 1 [1 G…
2 2006 9 2008 3001 NA NA 2 3 [BAL… 503 1 [1 G…
3 2006 9 2010 6001 NA(i) NA 2 3 [BAL… 508 1 [1 G…
4 2006 10 2010 6002 NA(i) NA 1 1 [BAL… 408 1 [1 G…
5 2006 10 2006 10 2 1957 2 1 [BAL… 630 2 [2 G…
6 2006 10 2008 3002 NA NA 2 1 [BAL… 426 2 [2 G…
7 2006 11 2008 3003 NA NA 2 3 [BAL… 718 4 [2 G…
8 2006 11 2010 6003 NA(i) NA NA(n) 3 [BAL… 518 2 [2 G…
9 2006 11 2006 11 2 1957 2 3 [BAL… 630 4 [2 G…
10 2006 12 2010 6004 NA(i) NA 4 1 [BAL… 324 2 [2 G…
# ℹ 14,600 more rows
# ℹ 2,747 more variables: form <dbl+lbl>, formwt <dbl>, gender1 <dbl+lbl>,
# hompop <dbl+lbl>, intage <dbl+lbl>, intid <dbl+lbl>, intyrs <dbl+lbl>,
# mode <dbl+lbl>, oversamp <dbl>, phase <dbl+lbl>, race <dbl+lbl>,
# reg16 <dbl+lbl>, region <dbl+lbl>, relate1 <dbl+lbl>, relhh1 <dbl+lbl>,
# relhhd1 <dbl+lbl>, respnum <dbl+lbl>, rvisitor <dbl+lbl>,
# sampcode <dbl+lbl>, sample <dbl+lbl>, sex <dbl+lbl>, size <dbl+lbl>, …The GSS panel
- Many variables.
- Stata’s missing value types are preserved
- Data types are things like
dbl+lblindicating that Stata’s numeric values and variable labels have been preserved.
The GSS panel
- You can see the labeling system at work:
The GSS panel
- Values get pivoted, not labels, though.
gss_panel |>
select(sex, degree) |>
group_by(sex, degree) |>
tally() |>
pivot_wider(names_from = sex, values_from = n)# A tibble: 6 × 3
degree `1` `2`
<dbl+lbl> <int> <int>
1 0 [LT HIGH SCHOOL] 814 1036
2 1 [HIGH SCHOOL] 3131 4143
3 2 [JUNIOR COLLEGE] 440 721
4 3 [bachelor] 1293 1474
5 4 [graduate] 696 860
6 NA(d) NA 2The GSS panel
- Option 1: Just drop all the labels.
# A tibble: 14,610 × 2,757
firstyear firstid year id vpsu vstrat adults ballot dateintv famgen
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 2006 9 2006 9 2 1957 1 3 709 1
2 2006 9 2008 3001 NA NA 2 3 503 1
3 2006 9 2010 6001 NA NA 2 3 508 1
4 2006 10 2010 6002 NA NA 1 1 408 1
5 2006 10 2006 10 2 1957 2 1 630 2
6 2006 10 2008 3002 NA NA 2 1 426 2
7 2006 11 2008 3003 NA NA 2 3 718 4
8 2006 11 2010 6003 NA NA NA 3 518 2
9 2006 11 2006 11 2 1957 2 3 630 4
10 2006 12 2010 6004 NA NA 4 1 324 2
# ℹ 14,600 more rows
# ℹ 2,747 more variables: form <dbl>, formwt <dbl>, gender1 <dbl>,
# hompop <dbl>, intage <dbl>, intid <dbl>, intyrs <dbl>, mode <dbl>,
# oversamp <dbl>, phase <dbl>, race <dbl>, reg16 <dbl>, region <dbl>,
# relate1 <dbl>, relhh1 <dbl>, relhhd1 <dbl>, respnum <dbl>, rvisitor <dbl>,
# sampcode <dbl>, sample <dbl>, sex <dbl>, size <dbl>, spaneng <dbl>,
# srcbelt <dbl>, version <dbl>, visitors <dbl>, wtss <dbl>, wtssall <dbl>, …The GSS panel
- Option 2: Convert the labels
- Let’s focus on a few measures of interest, and do some recoding.
## Categorical vars
cat_vars <- c("race", "sex", "degree", "relig", "income", "polviews", "fefam")
## Integer vars
int_vars <- c("year", "id", "ballot", "age", "tvhours")
## Survey design
wt_vars <- c("vpsu",
"vstrat",
"oversamp",
"formwt", # weight to deal with experimental randomization
"wtssall", # weight variable
"sampcode", # sampling error code
"sample") # sampling frame and method
my_gss_vars <- c(int_vars, cat_vars, wt_vars)Cut down the dataset
# A tibble: 14,610 × 19
year id ballot age tvhours race sex degree relig
<dbl> <dbl> <dbl+lbl> <dbl+lb> <dbl+lbl> <dbl+l> <dbl+l> <dbl+l> <dbl+l>
1 2006 9 3 [BALLOT C] 23 NA(a) [iap] 2 [bla… 2 [fem… 3 [bac… 4 [non…
2 2008 3001 3 [BALLOT C] 25 NA(i) 3 [oth… 2 [fem… 3 [bac… 4 [non…
3 2010 6001 3 [BALLOT C] 27 NA(i) 2 [bla… 2 [fem… 3 [bac… 4 [non…
4 2010 6002 1 [BALLOT A] 36 3 1 [whi… 2 [fem… 4 [gra… 4 [non…
5 2006 10 1 [BALLOT A] 32 3 3 [oth… 2 [fem… 4 [gra… 4 [non…
6 2008 3002 1 [BALLOT A] 34 3 3 [oth… 2 [fem… 4 [gra… 4 [non…
7 2008 3003 3 [BALLOT C] 83 NA(i) 2 [bla… 2 [fem… 0 [LT … 1 [pro…
8 2010 6003 3 [BALLOT C] 85 NA(i) 2 [bla… 2 [fem… 0 [LT … 1 [pro…
9 2006 11 3 [BALLOT C] 81 NA(a) [iap] 2 [bla… 2 [fem… 0 [LT … 1 [pro…
10 2010 6004 1 [BALLOT A] 51 10 3 [oth… 1 [mal… 1 [HIG… 2 [cat…
# ℹ 14,600 more rows
# ℹ 10 more variables: income <dbl+lbl>, polviews <dbl+lbl>, fefam <dbl+lbl>,
# vpsu <dbl+lbl>, vstrat <dbl+lbl>, oversamp <dbl>, formwt <dbl>,
# wtssall <dbl+lbl>, sampcode <dbl+lbl>, sample <dbl+lbl>The GSS Panel: Recoding
gss_sub |>
mutate(across(everything(), zap_missing)) |>
mutate(across(all_of(wt_vars), as.numeric)) |>
mutate(across(all_of(int_vars), as.integer)) |>
mutate(across(all_of(cat_vars), as_factor)) |>
mutate(across(all_of(cat_vars), fct_relabel, tolower)) |>
mutate(across(all_of(cat_vars), fct_relabel, tools::toTitleCase)) |>
mutate(income = str_replace(income, " - ", "-")) # A tibble: 14,610 × 19
year id ballot age tvhours race sex degree relig income polviews
<int> <int> <int> <int> <int> <fct> <fct> <fct> <fct> <chr> <fct>
1 2006 9 3 23 NA Black Female Bachelor None $2500… Conserv…
2 2008 3001 3 25 NA Other Female Bachelor None $2500… Extreme…
3 2010 6001 3 27 NA Black Female Bachelor None $2500… Extreme…
4 2010 6002 1 36 3 White Female Graduate None $2500… Liberal
5 2006 10 1 32 3 Other Female Graduate None <NA> Slightl…
6 2008 3002 1 34 3 Other Female Graduate None $2500… Moderate
7 2008 3003 3 83 NA Black Female Lt High … Prot… $2000… Liberal
8 2010 6003 3 85 NA Black Female Lt High … Prot… <NA> Moderate
9 2006 11 3 81 NA Black Female Lt High … Prot… <NA> Moderate
10 2010 6004 1 51 10 Other Male High Sch… Cath… Lt $1… Liberal
# ℹ 14,600 more rows
# ℹ 8 more variables: fefam <fct>, vpsu <dbl>, vstrat <dbl>, oversamp <dbl>,
# formwt <dbl>, wtssall <dbl>, sampcode <dbl>, sample <dbl>How we’d actually write this
gss_sub <- gss_sub |>
mutate(across(everything(), zap_missing),
across(all_of(wt_vars), as.numeric),
across(all_of(int_vars), as.integer),
across(all_of(cat_vars), as_factor),
across(all_of(cat_vars), fct_relabel, tolower),
across(all_of(cat_vars), fct_relabel, tools::toTitleCase),
income = str_replace(income, " - ", "-")) The GSS panel: more recoding
- Age quintiles: find the cutpoints
The GSS panel: more recoding
- Age quintiles: create the quintile variable
## Apply the cut
gss_sub |>
mutate(agequint = cut(x = age,
breaks = unique(age_quintiles),
include.lowest = TRUE)) |>
pull(agequint) |> # grab a column and make it an ordinary vector
table()
[18,33] (33,43] (43,53] (53,65] (65,89]
3157 2680 2851 3057 2720 - We’ll need to clean up those labels.
The GSS panel: more recoding
- I told you that regexp stuff would pay off.
convert_agegrp <- function(x){
x <- stringr::str_remove(x, "\\(") # Remove open paren
x <- stringr::str_remove(x, "\\[") # Remove open bracket
x <- stringr::str_remove(x, "\\]") # Remove close bracket
x <- stringr::str_replace(x, ",", "-") # Replace comma with dash
x <- stringr::str_replace(x, "-89", "+") # Replace -89 with +
regex <- "^(.*$)" # Matches everything in string to end of line
x <- stringr::str_replace(x, regex, "Age \\1") # Preface string with "Age"
x
}The GSS panel: more recoding
# A tibble: 14,610 × 19
year id ballot age tvhours race sex degree relig income polviews
<int> <int> <int> <int> <int> <fct> <fct> <fct> <fct> <chr> <fct>
1 2006 9 3 23 NA Black Female Bachelor None $2500… Conserv…
2 2008 3001 3 25 NA Other Female Bachelor None $2500… Extreme…
3 2010 6001 3 27 NA Black Female Bachelor None $2500… Extreme…
4 2010 6002 1 36 3 White Female Graduate None $2500… Liberal
5 2006 10 1 32 3 Other Female Graduate None <NA> Slightl…
6 2008 3002 1 34 3 Other Female Graduate None $2500… Moderate
7 2008 3003 3 83 NA Black Female Lt High … Prot… $2000… Liberal
8 2010 6003 3 85 NA Black Female Lt High … Prot… <NA> Moderate
9 2006 11 3 81 NA Black Female Lt High … Prot… <NA> Moderate
10 2010 6004 1 51 10 Other Male High Sch… Cath… Lt $1… Liberal
# ℹ 14,600 more rows
# ℹ 8 more variables: fefam <fct>, vpsu <dbl>, vstrat <dbl>, oversamp <dbl>,
# formwt <dbl>, wtssall <dbl>, sampcode <dbl>, sample <dbl>The GSS panel: more recoding
# A tibble: 14,610 × 4
age year degree fefam
<int> <int> <fct> <fct>
1 23 2006 Bachelor <NA>
2 25 2008 Bachelor <NA>
3 27 2010 Bachelor <NA>
4 36 2010 Graduate Disagree
5 32 2006 Graduate Agree
6 34 2008 Graduate Disagree
7 83 2008 Lt High School <NA>
8 85 2010 Lt High School <NA>
9 81 2006 Lt High School <NA>
10 51 2010 High School Disagree
# ℹ 14,600 more rowsThe GSS panel: more recoding
# A tibble: 14,610 × 5
age year degree fefam agequint
<int> <int> <fct> <fct> <fct>
1 23 2006 Bachelor <NA> [18,33]
2 25 2008 Bachelor <NA> [18,33]
3 27 2010 Bachelor <NA> [18,33]
4 36 2010 Graduate Disagree (33,43]
5 32 2006 Graduate Agree [18,33]
6 34 2008 Graduate Disagree (33,43]
7 83 2008 Lt High School <NA> (65,89]
8 85 2010 Lt High School <NA> (65,89]
9 81 2006 Lt High School <NA> (65,89]
10 51 2010 High School Disagree (43,53]
# ℹ 14,600 more rowsThe GSS panel: more recoding
# A tibble: 14,610 × 5
age year degree fefam agequint
<int> <int> <fct> <fct> <fct>
1 23 2006 Bachelor <NA> Age 18-33
2 25 2008 Bachelor <NA> Age 18-33
3 27 2010 Bachelor <NA> Age 18-33
4 36 2010 Graduate Disagree Age 33-43
5 32 2006 Graduate Agree Age 18-33
6 34 2008 Graduate Disagree Age 33-43
7 83 2008 Lt High School <NA> Age 65+
8 85 2010 Lt High School <NA> Age 65+
9 81 2006 Lt High School <NA> Age 65+
10 51 2010 High School Disagree Age 43-53
# ℹ 14,600 more rowsThe GSS panel: more recoding
gss_sub |>
# `select` is here just so you can see
# what's happening to the columns
select(age, year, degree, fefam) |>
mutate(agequint = cut(x = age,
breaks = unique(age_quintiles),
include.lowest = TRUE)) |>
mutate(agequint = fct_relabel(agequint, convert_agegrp)) |>
mutate(year_f = droplevels(factor(year)))# A tibble: 14,610 × 6
age year degree fefam agequint year_f
<int> <int> <fct> <fct> <fct> <fct>
1 23 2006 Bachelor <NA> Age 18-33 2006
2 25 2008 Bachelor <NA> Age 18-33 2008
3 27 2010 Bachelor <NA> Age 18-33 2010
4 36 2010 Graduate Disagree Age 33-43 2010
5 32 2006 Graduate Agree Age 18-33 2006
6 34 2008 Graduate Disagree Age 33-43 2008
7 83 2008 Lt High School <NA> Age 65+ 2008
8 85 2010 Lt High School <NA> Age 65+ 2010
9 81 2006 Lt High School <NA> Age 65+ 2006
10 51 2010 High School Disagree Age 43-53 2010
# ℹ 14,600 more rowsThe GSS panel: more recoding
gss_sub |>
# `select` is here just so you can see
# what's happening to the columns
select(age, year, degree, fefam) |>
mutate(agequint = cut(x = age,
breaks = unique(age_quintiles),
include.lowest = TRUE)) |>
mutate(agequint = fct_relabel(agequint, convert_agegrp)) |>
mutate(year_f = droplevels(factor(year))) |>
mutate(young = ifelse(age < 26, "Yes", "No"))# A tibble: 14,610 × 7
age year degree fefam agequint year_f young
<int> <int> <fct> <fct> <fct> <fct> <chr>
1 23 2006 Bachelor <NA> Age 18-33 2006 Yes
2 25 2008 Bachelor <NA> Age 18-33 2008 Yes
3 27 2010 Bachelor <NA> Age 18-33 2010 No
4 36 2010 Graduate Disagree Age 33-43 2010 No
5 32 2006 Graduate Agree Age 18-33 2006 No
6 34 2008 Graduate Disagree Age 33-43 2008 No
7 83 2008 Lt High School <NA> Age 65+ 2008 No
8 85 2010 Lt High School <NA> Age 65+ 2010 No
9 81 2006 Lt High School <NA> Age 65+ 2006 No
10 51 2010 High School Disagree Age 43-53 2010 No
# ℹ 14,600 more rowsThe GSS panel: more recoding
gss_sub |>
# `select` is here just so you can see
# what's happening to the columns
select(age, year, degree, fefam) |>
mutate(agequint = cut(x = age,
breaks = unique(age_quintiles),
include.lowest = TRUE)) |>
mutate(agequint = fct_relabel(agequint, convert_agegrp)) |>
mutate(year_f = droplevels(factor(year))) |>
mutate(young = ifelse(age < 26, "Yes", "No")) |>
mutate(fefam_d = fct_recode(fefam,
Agree = "Strongly Agree",
Disagree = "Strongly Disagree"))# A tibble: 14,610 × 8
age year degree fefam agequint year_f young fefam_d
<int> <int> <fct> <fct> <fct> <fct> <chr> <fct>
1 23 2006 Bachelor <NA> Age 18-33 2006 Yes <NA>
2 25 2008 Bachelor <NA> Age 18-33 2008 Yes <NA>
3 27 2010 Bachelor <NA> Age 18-33 2010 No <NA>
4 36 2010 Graduate Disagree Age 33-43 2010 No Disagree
5 32 2006 Graduate Agree Age 18-33 2006 No Agree
6 34 2008 Graduate Disagree Age 33-43 2008 No Disagree
7 83 2008 Lt High School <NA> Age 65+ 2008 No <NA>
8 85 2010 Lt High School <NA> Age 65+ 2010 No <NA>
9 81 2006 Lt High School <NA> Age 65+ 2006 No <NA>
10 51 2010 High School Disagree Age 43-53 2010 No Disagree
# ℹ 14,600 more rowsThe GSS panel: more recoding
gss_sub |>
# `select` is here just so you can see
# what's happening to the columns
select(age, year, degree, fefam) |>
mutate(agequint = cut(x = age,
breaks = unique(age_quintiles),
include.lowest = TRUE)) |>
mutate(agequint = fct_relabel(agequint, convert_agegrp)) |>
mutate(year_f = droplevels(factor(year))) |>
mutate(young = ifelse(age < 26, "Yes", "No")) |>
mutate(fefam_d = fct_recode(fefam,
Agree = "Strongly Agree",
Disagree = "Strongly Disagree")) |>
mutate(degree = factor(degree,
levels = levels(gss_sub$degree),
ordered = TRUE)) # A tibble: 14,610 × 8
age year degree fefam agequint year_f young fefam_d
<int> <int> <ord> <fct> <fct> <fct> <chr> <fct>
1 23 2006 Bachelor <NA> Age 18-33 2006 Yes <NA>
2 25 2008 Bachelor <NA> Age 18-33 2008 Yes <NA>
3 27 2010 Bachelor <NA> Age 18-33 2010 No <NA>
4 36 2010 Graduate Disagree Age 33-43 2010 No Disagree
5 32 2006 Graduate Agree Age 18-33 2006 No Agree
6 34 2008 Graduate Disagree Age 33-43 2008 No Disagree
7 83 2008 Lt High School <NA> Age 65+ 2008 No <NA>
8 85 2010 Lt High School <NA> Age 65+ 2010 No <NA>
9 81 2006 Lt High School <NA> Age 65+ 2006 No <NA>
10 51 2010 High School Disagree Age 43-53 2010 No Disagree
# ℹ 14,600 more rowsHow we’d actually write this
gss_sub <- gss_sub |>
mutate(agequint = cut(x = age,
breaks = unique(age_quintiles),
include.lowest = TRUE),
agequint = fct_relabel(agequint, convert_agegrp),
year_f = droplevels(factor(year)),
young = ifelse(age < 26, "Yes", "No"),
fefam_d = fct_recode(fefam,
Agree = "Strongly Agree",
Disagree = "Strongly Disagree"),
degree = factor(degree,
levels = levels(gss_sub$degree),
ordered = TRUE))
gss_sub# A tibble: 14,610 × 23
year id ballot age tvhours race sex degree relig income polviews
<int> <int> <int> <int> <int> <fct> <fct> <ord> <fct> <chr> <fct>
1 2006 9 3 23 NA Black Female Bachelor None $2500… Conserv…
2 2008 3001 3 25 NA Other Female Bachelor None $2500… Extreme…
3 2010 6001 3 27 NA Black Female Bachelor None $2500… Extreme…
4 2010 6002 1 36 3 White Female Graduate None $2500… Liberal
5 2006 10 1 32 3 Other Female Graduate None <NA> Slightl…
6 2008 3002 1 34 3 Other Female Graduate None $2500… Moderate
7 2008 3003 3 83 NA Black Female Lt High … Prot… $2000… Liberal
8 2010 6003 3 85 NA Black Female Lt High … Prot… <NA> Moderate
9 2006 11 3 81 NA Black Female Lt High … Prot… <NA> Moderate
10 2010 6004 1 51 10 Other Male High Sch… Cath… Lt $1… Liberal
# ℹ 14,600 more rows
# ℹ 12 more variables: fefam <fct>, vpsu <dbl>, vstrat <dbl>, oversamp <dbl>,
# formwt <dbl>, wtssall <dbl>, sampcode <dbl>, sample <dbl>, agequint <fct>,
# year_f <fct>, young <chr>, fefam_d <fct>How we’d actually write this
gss_sub <- gss_sub |>
mutate(agequint = cut(x = age,
breaks = unique(age_quintiles),
include.lowest = TRUE),
agequint = fct_relabel(agequint, convert_agegrp),
year_f = factor(year),
young = ifelse(age < 26, "Yes", "No"),
fefam_d = fct_recode(fefam,
Agree = "Strongly Agree",
Disagree = "Strongly Disagree"),
degree = factor(degree,
levels = levels(gss_sub$degree),
ordered = TRUE))How we’d actually write this
gss_sub <- gss_sub |>
mutate(agequint = cut(x = age,
breaks = unique(age_quintiles),
include.lowest = TRUE),
agequint = fct_relabel(agequint, convert_agegrp),
year_f = droplevels(factor(year)),
young = ifelse(age < 26, "Yes", "No"),
fefam_d = fct_recode(fefam,
Agree = "Strongly Agree",
Disagree = "Strongly Disagree"),
degree = factor(degree,
levels = levels(gss_sub$degree),
ordered = TRUE))GSS Panel
# A tibble: 15 × 7
sex year year_f age young fefam fefam_d
<fct> <int> <fct> <int> <chr> <fct> <fct>
1 Female 2010 2010 46 No Disagree Disagree
2 Female 2012 2012 74 No Agree Agree
3 Male 2010 2010 48 No Disagree Disagree
4 Male 2008 2008 23 Yes Agree Agree
5 Male 2008 2008 75 No Strongly Agree Agree
6 Male 2010 2010 54 No <NA> <NA>
7 Female 2010 2010 60 No Strongly Disagree Disagree
8 Female 2012 2012 41 No <NA> <NA>
9 Female 2008 2008 75 No Agree Agree
10 Female 2008 2008 76 No <NA> <NA>
11 Male 2010 2010 39 No Disagree Disagree
12 Female 2010 2010 71 No Agree Agree
13 Male 2010 2010 59 No Agree Agree
14 Female 2010 2010 55 No <NA> <NA>
15 Male 2010 2010 42 No <NA> <NA> GSS Panel
More about factors
More on factors
- We’ve already seen
fct_relabel()andfct_recode()from forcats. - There are numerous other convenience functions for factors.
More on factors
# A tibble: 6 × 2
degree n
<ord> <int>
1 Lt High School 1850
2 High School 7274
3 Junior College 1161
4 Bachelor 2767
5 Graduate 1556
6 <NA> 2More on factors
- Make the NA values an explicit level
More on factors
- Relevel by frequency
More on factors
- Relevel manually
More on factors
- Relevel manually
Call:
lm(formula = age ~ sex, data = gss_sub)
Residuals:
Min 1Q Median 3Q Max
-31.431 -13.972 -0.431 12.569 40.028
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 48.9720 0.2149 227.846 <2e-16 ***
sexFemale 0.4594 0.2864 1.604 0.109
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 17.08 on 14463 degrees of freedom
(145 observations deleted due to missingness)
Multiple R-squared: 0.0001779, Adjusted R-squared: 0.0001088
F-statistic: 2.573 on 1 and 14463 DF, p-value: 0.1087More on factors
- Relevel manually
More on factors
- Relevel manually
Call:
lm(formula = age ~ sex, data = gss_sub)
Residuals:
Min 1Q Median 3Q Max
-31.431 -13.972 -0.431 12.569 40.028
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 49.4313 0.1892 261.233 <2e-16 ***
sexMale -0.4594 0.2864 -1.604 0.109
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 17.08 on 14463 degrees of freedom
(145 observations deleted due to missingness)
Multiple R-squared: 0.0001779, Adjusted R-squared: 0.0001088
F-statistic: 2.573 on 1 and 14463 DF, p-value: 0.1087More on factors
- Interact or cross factors
gss_sub <- gss_sub |>
mutate(degree_by_race = fct_cross(race, degree))
gss_sub |>
count(degree_by_race)# A tibble: 16 × 2
degree_by_race n
<fct> <int>
1 White:Lt High School 1188
2 Black:Lt High School 379
3 Other:Lt High School 283
4 White:High School 5548
5 Black:High School 1180
6 Other:High School 546
7 White:Junior College 885
8 Black:Junior College 206
9 Other:Junior College 70
10 White:Bachelor 2334
11 Black:Bachelor 233
12 Other:Bachelor 200
13 White:Graduate 1293
14 Black:Graduate 116
15 Other:Graduate 147
16 <NA> 2More on factors
- Relevel manually by lumping … the least frequent n
More on factors
Relevel manually by lumping …to other, manually