library(here) # manage file paths
library(socviz) # data and some useful functions
library(tidyverse) # your friend and mineHelpers, Build Systems, Environments, Packages
Modern Plain Text Social Science: Week 12
Kieran Healy
Duke University
November 26, 2024
Helpers, Build Systems, Environments, and Packages
Load the packages, as always
Helpers
The RStudio Community
The reprex package
Best demonstrated live
When asking for help, make a reproducible example
library(tidyverse)
starwars |>
count(homeworld, species) |>
mutate(pct = n / sum(n) * 100) |>
arrange(desc(pct))# A tibble: 57 × 4
homeworld species n pct
<chr> <chr> <int> <dbl>
1 Tatooine Human 8 9.20
2 <NA> Human 6 6.90
3 Naboo Human 5 5.75
4 Alderaan Human 3 3.45
5 Naboo Gungan 3 3.45
6 <NA> Droid 3 3.45
7 Corellia Human 2 2.30
8 Coruscant Human 2 2.30
9 Kamino Kaminoan 2 2.30
10 Kashyyyk Wookiee 2 2.30
# ℹ 47 more rowsThe usethis package
Tables, tables, tables
tinytable
| mpg | cyl | disp | hp | drat | wt | qsec | vs | am | gear | carb |
|---|---|---|---|---|---|---|---|---|---|---|
| 21.0 | 6 | 160.0 | 110 | 3.90 | 2.620 | 16.46 | 0 | 1 | 4 | 4 |
| 21.0 | 6 | 160.0 | 110 | 3.90 | 2.875 | 17.02 | 0 | 1 | 4 | 4 |
| 22.8 | 4 | 108.0 | 93 | 3.85 | 2.320 | 18.61 | 1 | 1 | 4 | 1 |
| 21.4 | 6 | 258.0 | 110 | 3.08 | 3.215 | 19.44 | 1 | 0 | 3 | 1 |
| 18.7 | 8 | 360.0 | 175 | 3.15 | 3.440 | 17.02 | 0 | 0 | 3 | 2 |
| 18.1 | 6 | 225.0 | 105 | 2.76 | 3.460 | 20.22 | 1 | 0 | 3 | 1 |
| 14.3 | 8 | 360.0 | 245 | 3.21 | 3.570 | 15.84 | 0 | 0 | 3 | 4 |
| 24.4 | 4 | 146.7 | 62 | 3.69 | 3.190 | 20.00 | 1 | 0 | 4 | 2 |
| 22.8 | 4 | 140.8 | 95 | 3.92 | 3.150 | 22.90 | 1 | 0 | 4 | 2 |
| 19.2 | 6 | 167.6 | 123 | 3.92 | 3.440 | 18.30 | 1 | 0 | 4 | 4 |
| 17.8 | 6 | 167.6 | 123 | 3.92 | 3.440 | 18.90 | 1 | 0 | 4 | 4 |
| 16.4 | 8 | 275.8 | 180 | 3.07 | 4.070 | 17.40 | 0 | 0 | 3 | 3 |
| 17.3 | 8 | 275.8 | 180 | 3.07 | 3.730 | 17.60 | 0 | 0 | 3 | 3 |
| 15.2 | 8 | 275.8 | 180 | 3.07 | 3.780 | 18.00 | 0 | 0 | 3 | 3 |
| 10.4 | 8 | 472.0 | 205 | 2.93 | 5.250 | 17.98 | 0 | 0 | 3 | 4 |
| 10.4 | 8 | 460.0 | 215 | 3.00 | 5.424 | 17.82 | 0 | 0 | 3 | 4 |
| 14.7 | 8 | 440.0 | 230 | 3.23 | 5.345 | 17.42 | 0 | 0 | 3 | 4 |
| 32.4 | 4 | 78.7 | 66 | 4.08 | 2.200 | 19.47 | 1 | 1 | 4 | 1 |
| 30.4 | 4 | 75.7 | 52 | 4.93 | 1.615 | 18.52 | 1 | 1 | 4 | 2 |
| 33.9 | 4 | 71.1 | 65 | 4.22 | 1.835 | 19.90 | 1 | 1 | 4 | 1 |
| 21.5 | 4 | 120.1 | 97 | 3.70 | 2.465 | 20.01 | 1 | 0 | 3 | 1 |
| 15.5 | 8 | 318.0 | 150 | 2.76 | 3.520 | 16.87 | 0 | 0 | 3 | 2 |
| 15.2 | 8 | 304.0 | 150 | 3.15 | 3.435 | 17.30 | 0 | 0 | 3 | 2 |
| 13.3 | 8 | 350.0 | 245 | 3.73 | 3.840 | 15.41 | 0 | 0 | 3 | 4 |
| 19.2 | 8 | 400.0 | 175 | 3.08 | 3.845 | 17.05 | 0 | 0 | 3 | 2 |
| 27.3 | 4 | 79.0 | 66 | 4.08 | 1.935 | 18.90 | 1 | 1 | 4 | 1 |
| 26.0 | 4 | 120.3 | 91 | 4.43 | 2.140 | 16.70 | 0 | 1 | 5 | 2 |
| 30.4 | 4 | 95.1 | 113 | 3.77 | 1.513 | 16.90 | 1 | 1 | 5 | 2 |
| 15.8 | 8 | 351.0 | 264 | 4.22 | 3.170 | 14.50 | 0 | 1 | 5 | 4 |
| 19.7 | 6 | 145.0 | 175 | 3.62 | 2.770 | 15.50 | 0 | 1 | 5 | 6 |
| 15.0 | 8 | 301.0 | 335 | 3.54 | 3.570 | 14.60 | 0 | 1 | 5 | 8 |
| 21.4 | 4 | 121.0 | 109 | 4.11 | 2.780 | 18.60 | 1 | 1 | 4 | 2 |
gt
From its website:
library(gt)
# Define the start and end dates for the data range
start_date <- "2010-06-07"
end_date <- "2010-06-14"
# Create a gt table based on preprocessed
# `sp500` table data
sp500 |>
dplyr::filter(date >= start_date & date <= end_date) |>
dplyr::select(-adj_close) |>
gt() |>
tab_header(
title = "S&P 500",
subtitle = glue::glue("{start_date} to {end_date}")
) |>
fmt_currency() |>
fmt_date(columns = date, date_style = "wd_m_day_year") |>
fmt_number(columns = volume, suffixing = TRUE)| S&P 500 | |||||
|---|---|---|---|---|---|
| 2010-06-07 to 2010-06-14 | |||||
| date | open | high | low | close | volume |
| Mon, Jun 14, 2010 | $1,095.00 | $1,105.91 | $1,089.03 | $1,089.63 | 4.43B |
| Fri, Jun 11, 2010 | $1,082.65 | $1,092.25 | $1,077.12 | $1,091.60 | 4.06B |
| Thu, Jun 10, 2010 | $1,058.77 | $1,087.85 | $1,058.77 | $1,086.84 | 5.14B |
| Wed, Jun 9, 2010 | $1,062.75 | $1,077.74 | $1,052.25 | $1,055.69 | 5.98B |
| Tue, Jun 8, 2010 | $1,050.81 | $1,063.15 | $1,042.17 | $1,062.00 | 6.19B |
| Mon, Jun 7, 2010 | $1,065.84 | $1,071.36 | $1,049.86 | $1,050.47 | 5.47B |
gtsummary
Built on top of gt, the gtsummary package.
# A tibble: 200 × 8
trt age marker stage grade response death ttdeath
<chr> <dbl> <dbl> <fct> <fct> <int> <int> <dbl>
1 Drug A 23 0.16 T1 II 0 0 24
2 Drug B 9 1.11 T2 I 1 0 24
3 Drug A 31 0.277 T1 II 0 0 24
4 Drug A NA 2.07 T3 III 1 1 17.6
5 Drug A 51 2.77 T4 III 1 1 16.4
6 Drug B 39 0.613 T4 I 0 1 15.6
7 Drug A 37 0.354 T1 II 0 0 24
8 Drug A 32 1.74 T1 I 0 1 18.4
9 Drug A 31 0.144 T1 II 0 0 24
10 Drug B 34 0.205 T3 I 0 1 10.5
# ℹ 190 more rowsgtsummary
Built on top of gt, the gtsummary package.
gtsummary
| Characteristic | N | Drug A N = 981 |
Drug B N = 1021 |
p-value |
|---|---|---|---|---|
| Age | 189 | 46 (37, 60) | 48 (39, 56) | |
| Grade | 200 | |||
| I | 35 (36%) | 33 (32%) | ||
| II | 32 (33%) | 36 (35%) | ||
| III | 31 (32%) | 33 (32%) | ||
| Tumor Response | 193 | 28 (29%) | 33 (34%) | |
| 1 Median (Q1, Q3); n (%) | ||||
gtsummary
gtsummary() straight out of the box:
gtsummary
| Characteristic | White N = 2,0961 |
Black N = 4871 |
Other N = 2761 |
|---|---|---|---|
| degree | |||
| Lt High School | 197 (9.4%) | 60 (12%) | 71 (26%) |
| High School | 1,057 (50%) | 292 (60%) | 112 (41%) |
| Junior College | 166 (7.9%) | 33 (6.8%) | 17 (6.2%) |
| Bachelor | 426 (20%) | 71 (15%) | 39 (14%) |
| Graduate | 250 (12%) | 31 (6.4%) | 37 (13%) |
| marital | |||
| Married | 979 (47%) | 121 (25%) | 110 (40%) |
| Widowed | 196 (9.4%) | 35 (7.2%) | 18 (6.5%) |
| Divorced | 363 (17%) | 93 (19%) | 39 (14%) |
| Separated | 55 (2.6%) | 27 (5.5%) | 20 (7.2%) |
| Never Married | 503 (24%) | 211 (43%) | 89 (32%) |
| 1 n (%) | |||
gtsummary
With a bit more work …
gtsummary
| Characteristic | Drug A N = 98 |
Drug B N = 102 |
|---|---|---|
| Age | ||
| N Non-missing | 91 | 98 |
| Mean (SD) | 47 (15) | 47 (14) |
| Median (Q1, Q3) | 46 (37, 60) | 48 (39, 56) |
| Min, Max | 6, 78 | 9, 83 |
| Marker Level (ng/mL) | ||
| N Non-missing | 92 | 98 |
| Mean (SD) | 1.02 (0.89) | 0.82 (0.83) |
| Median (Q1, Q3) | 0.84 (0.23, 1.60) | 0.52 (0.18, 1.21) |
| Min, Max | 0.00, 3.87 | 0.01, 3.64 |
Summarize with skimr
- We might want to make a codebook of our data
Summarize with skimr
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| year | 34 | 0.86 | 1996.50 | 3.46 | 1991.00 | 1993.75 | 1996.50 | 1999.25 | 2002.00 | ▇▅▅▅▇ |
| donors | 34 | 0.86 | 16.48 | 5.11 | 5.20 | 13.00 | 15.10 | 19.60 | 33.90 | ▁▇▅▂▁ |
| pop | 17 | 0.93 | 39921.29 | 62219.22 | 3514.00 | 6938.00 | 15531.00 | 57301.00 | 288369.00 | ▇▁▁▁▁ |
| pop.dens | 17 | 0.93 | 12.00 | 11.09 | 0.22 | 1.94 | 9.49 | 19.11 | 38.89 | ▇▃▃▂▁ |
| gdp | 17 | 0.93 | 22986.18 | 4665.92 | 12917.00 | 19546.00 | 22756.00 | 26180.00 | 36554.00 | ▂▇▇▃▁ |
| gdp.lag | 0 | 1.00 | 22574.92 | 4790.71 | 11434.00 | 19034.25 | 22158.00 | 25886.50 | 36554.00 | ▂▇▇▃▁ |
| health | 0 | 1.00 | 2073.75 | 733.59 | 791.00 | 1581.00 | 1956.00 | 2407.50 | 5665.00 | ▆▇▂▁▁ |
| health.lag | 0 | 1.00 | 1972.99 | 699.24 | 727.00 | 1542.00 | 1850.50 | 2290.25 | 5267.00 | ▆▇▂▁▁ |
| pubhealth | 21 | 0.91 | 6.19 | 0.92 | 4.30 | 5.50 | 6.00 | 6.90 | 8.80 | ▂▇▅▃▁ |
| roads | 17 | 0.93 | 113.04 | 36.33 | 58.21 | 83.46 | 111.22 | 139.57 | 232.48 | ▇▇▆▂▁ |
| cerebvas | 17 | 0.93 | 610.80 | 144.45 | 300.00 | 500.00 | 604.00 | 698.00 | 957.00 | ▂▅▇▃▂ |
| assault | 17 | 0.93 | 16.53 | 17.33 | 4.00 | 9.00 | 11.00 | 16.00 | 103.00 | ▇▁▁▁▁ |
| external | 17 | 0.93 | 450.06 | 118.19 | 258.00 | 367.00 | 421.00 | 534.00 | 853.00 | ▆▇▅▁▁ |
| txp.pop | 17 | 0.93 | 0.72 | 0.20 | 0.22 | 0.63 | 0.71 | 0.83 | 1.12 | ▁▂▇▃▃ |
Summarize with skimr
Variable type: character
| skim_variable | n_missing | complete_rate | min | max | empty | n_unique | whitespace |
|---|---|---|---|---|---|---|---|
| country | 0 | 1.00 | 5 | 14 | 0 | 17 | 0 |
| world | 14 | 0.94 | 6 | 11 | 0 | 3 | 0 |
| opt | 28 | 0.88 | 2 | 3 | 0 | 2 | 0 |
| consent.law | 0 | 1.00 | 8 | 8 | 0 | 2 | 0 |
| consent.practice | 0 | 1.00 | 8 | 8 | 0 | 2 | 0 |
| consistent | 0 | 1.00 | 2 | 3 | 0 | 2 | 0 |
| ccode | 0 | 1.00 | 2 | 4 | 0 | 17 | 0 |
Custom Summaries
# A tibble: 580,395 × 17
country_code cname iso2 continent iso3 year week sex split split_sex
<chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <chr> <dbl> <dbl>
1 AUS Austral… AU Oceania AUS 2015 1 m 1 0
2 AUS Austral… AU Oceania AUS 2015 1 m 1 0
3 AUS Austral… AU Oceania AUS 2015 1 m 1 0
4 AUS Austral… AU Oceania AUS 2015 1 m 1 0
5 AUS Austral… AU Oceania AUS 2015 1 m 1 0
6 AUS Austral… AU Oceania AUS 2015 1 f 1 0
7 AUS Austral… AU Oceania AUS 2015 1 f 1 0
8 AUS Austral… AU Oceania AUS 2015 1 f 1 0
9 AUS Austral… AU Oceania AUS 2015 1 f 1 0
10 AUS Austral… AU Oceania AUS 2015 1 f 1 0
# ℹ 580,385 more rows
# ℹ 7 more variables: forecast <dbl>, approx_date <date>, age_group <chr>,
# death_count <dbl>, death_rate <dbl>, deaths_total <dbl>, rate_total <dbl>Custom Summaries
stmf_country_years <- function(df = stmf) {
df |>
dplyr::select(cname, year) |>
dplyr::group_by(cname, year) |>
dplyr::tally() |>
dplyr::mutate(n = as.character(n),
n = dplyr::recode(n, "0" = "-", .default = "Y")) |>
dplyr::group_by(year, cname) |>
dplyr::arrange(year) |>
tidyr::pivot_wider(names_from = year, values_from = n) |>
dplyr::mutate(dplyr::across(where(is.character), \(x) dplyr::recode(x, .missing = "-"))) |>
dplyr::arrange(cname)
}Custom Summaries
| cname | 1990 | 1991 | 1992 | 1993 | 1994 | 1995 | 1996 | 1997 | 1998 | 1999 | 2000 | 2001 | 2002 | 2003 | 2004 | 2005 | 2006 | 2007 | 2008 | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | 2015 | 2016 | 2017 | 2018 | 2019 | 2020 | 2021 | 2022 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Australia | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y |
| Austria | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Belgium | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Bulgaria | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Canada | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Chile | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y |
| Croatia | - | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Czech Republic | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Denmark | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| England and Wales | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Estonia | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Finland | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| France | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Germany | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Greece | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y |
| Hungary | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Iceland | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Israel | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Italy | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Korea, Republic of | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Latvia | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Lithuania | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Luxembourg | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Netherlands | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| New Zealand | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Northern Ireland | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y |
| Norway | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Poland | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Portugal | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Russian Federation | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | - | - |
| Scotland | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Slovakia | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Slovenia | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Spain | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Sweden | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Switzerland | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Taiwan, Province of China | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | - |
| United States | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | Y | Y | Y | Y | Y | Y | Y |
How Projects Grow
Like Topsy
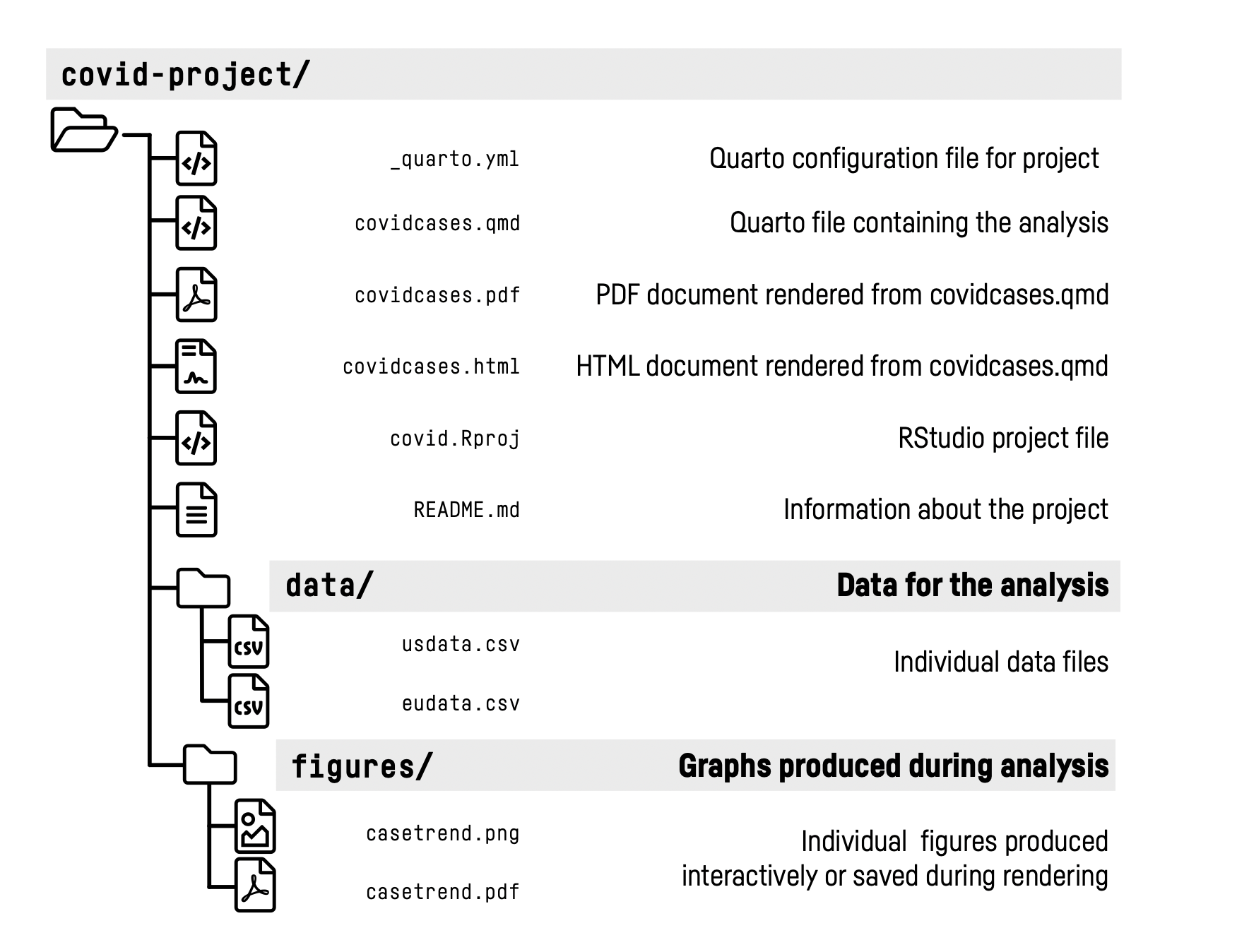
Like Topsy
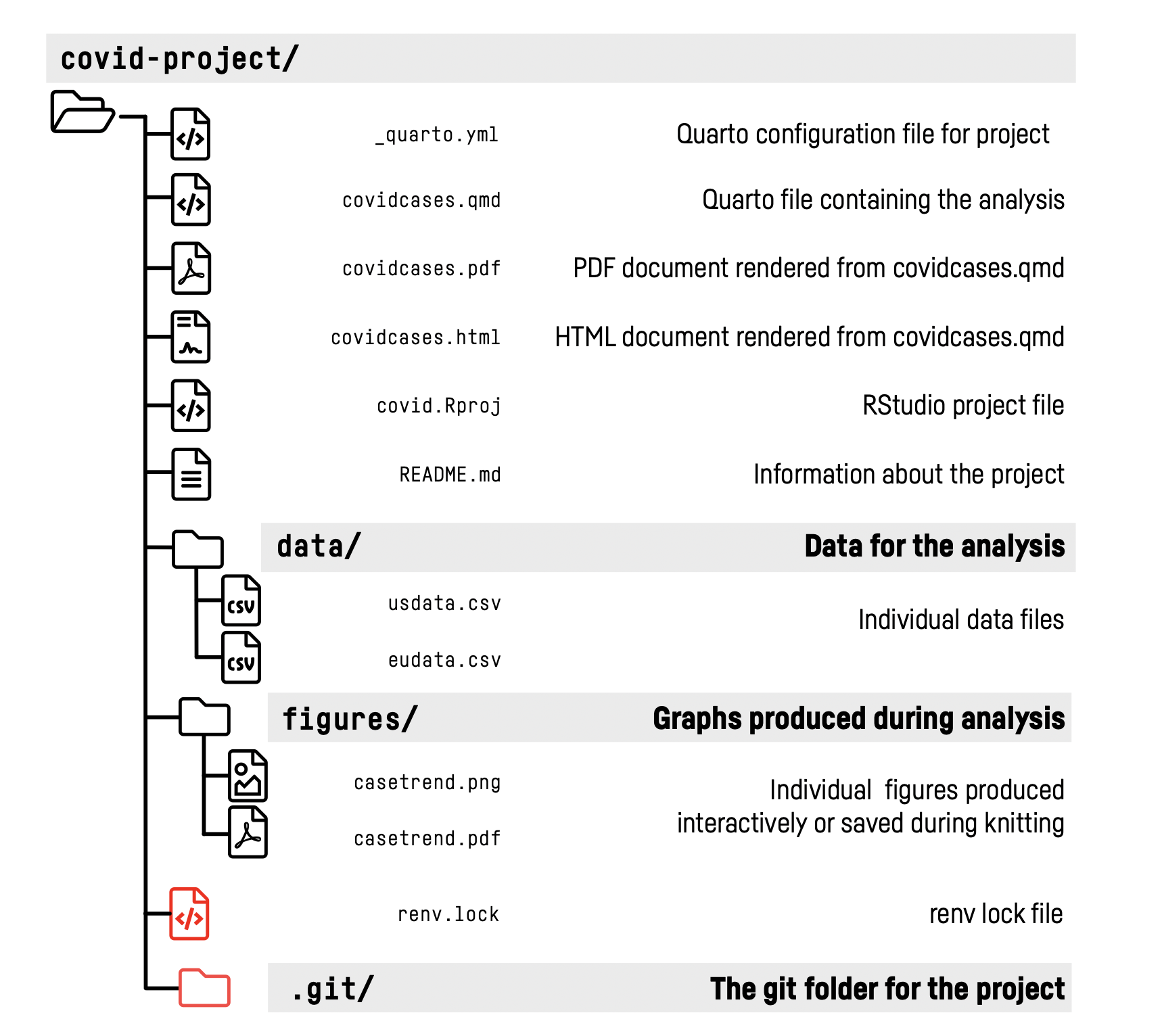
Like Topsy
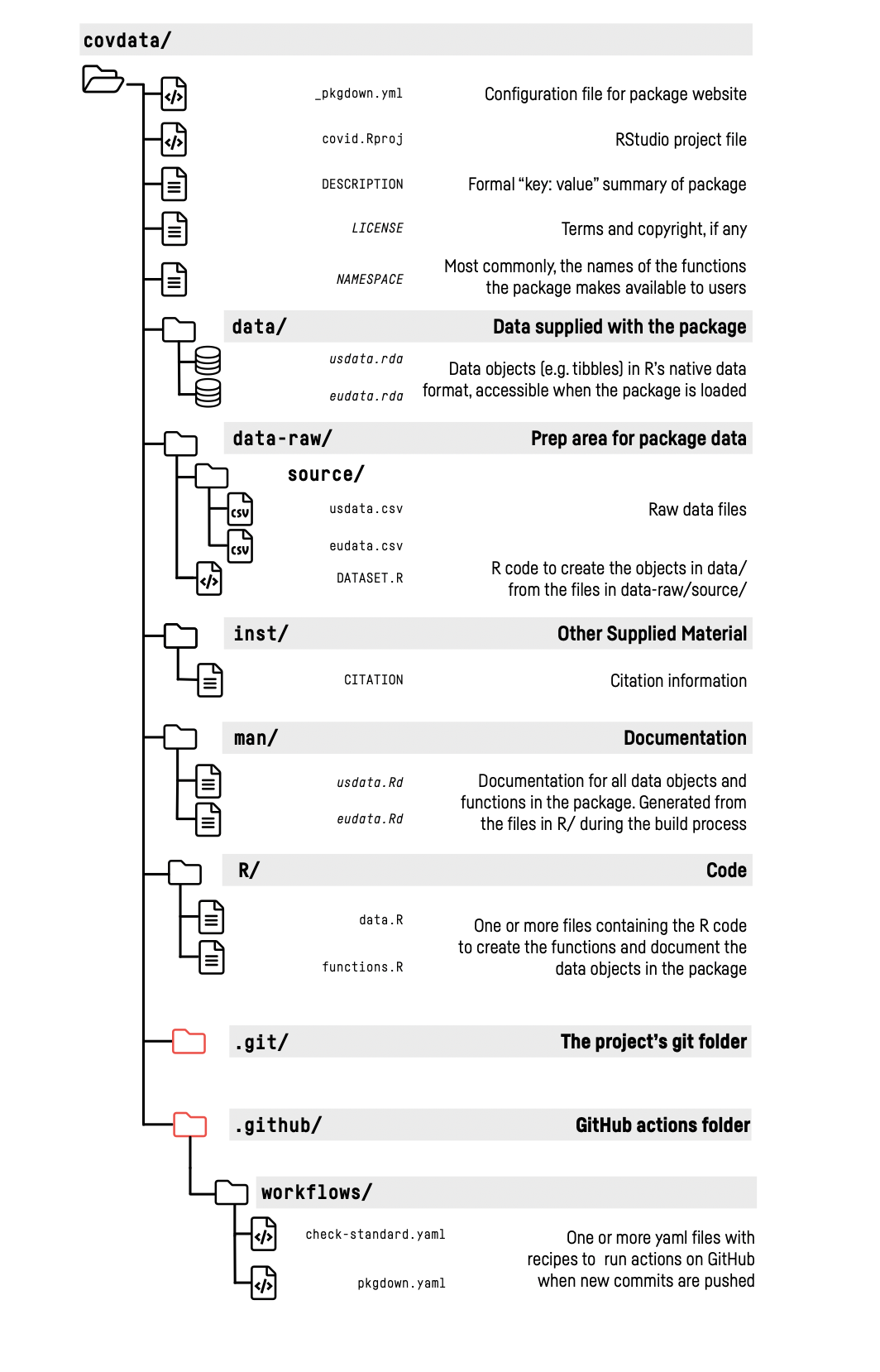
Controlling this
- As projects grow, you will want to refactor them
- Think in terms of modular code and invidivually isolatable steps
- These things tend to go in a predictable sequence
For instance
- Some scripts, or a single notebook, becomes …
- One script (or notebook) to rule them all, becomes …
- A coordinating script to rule them all, becomes …
- A function to rule them all, becomes …
- A series of functions to rule them all, becomes …
- A package to rule them all, becomes …
- A modular set of packages
Remember, everything is a DAG
Makefiles
Targets
Make for R: https://docs.ropensci.org/targets/
Comes with a very thorough manual: https://books.ropensci.org/targets/
renv and Build Environments
https://rstudio.github.io/renv/articles/renv.html
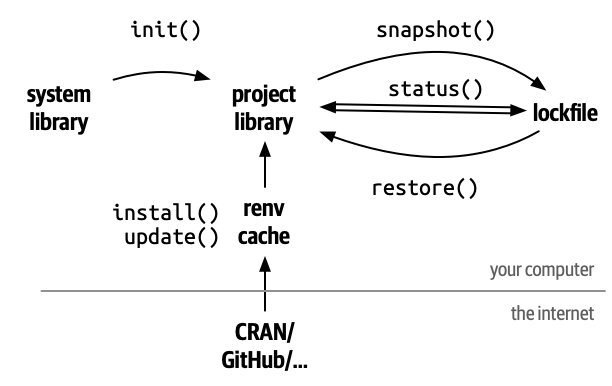
You check-in the lockfile to your project, but not the actual packages.
R Packages
For functionality, data, and papers
- Consider packaging your datasets! Benefits to documentation/codebooks etc
- One-table example: uscenpops
- More extensive: covdata, gssr
- How R packages work: Wickham & Bryan
Perspective
Solve the problem you have