── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.5.1 ✔ tibble 3.2.1
✔ lubridate 1.9.3 ✔ tidyr 1.3.1
✔ purrr 1.0.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
here() starts at /Users/kjhealy/Documents/courses/mptc
Loading required package: sysfonts
Loading required package: showtextdbExample 09: Functions and Iteration
library(broom)Functions
my_adder <- function(x, add = 1) {
x + add
}
my_adder(10)[1] 11my_adder(1:10) [1] 2 3 4 5 6 7 8 9 10 11# This won't work
# my_adder("a")Guard clauses and error handling:
my_adder <- function(x, add = 1) {
if(!is.numeric(x)) {
return("Argument `x` must be numeric.")
}
x + add
}
my_adder("a")[1] "Argument `x` must be numeric."my_adder <- function(x, add = 1) {
if(!is.numeric(x)) {
return("Argument `x` must be numeric.")
}
if(!is.numeric(add)) {
return("Argument `add` must be numeric.")
}
x + add
}Here I use return() in the guard clauses in order to make this example work as part of the pipeline that creates the site. But really I should use stop().
my_adder(1:10, 5) [1] 6 7 8 9 10 11 12 13 14 15my_adder(1:10, "c")[1] "Argument `add` must be numeric."Elvis PCA Example
An iconic image

Long Tall Sally
# install.packages("imager")
img <- imager::load.image(here("assets", "08-iterate", "elvis-nixon.jpeg"))
str(img) 'cimg' num [1:800, 1:633, 1, 1] 0.914 0.929 0.91 0.906 0.898 ...dim(img)[1] 800 633 1 1## Long
img_df_long <- as.data.frame(img)
head(img_df_long) x y value
1 1 1 0.9137255
2 2 1 0.9294118
3 3 1 0.9098039
4 4 1 0.9058824
5 5 1 0.8980392
6 6 1 0.8862745Return to Sender
img_df <- pivot_wider(img_df_long,
names_from = y,
values_from = value)
dim(img_df)[1] 800 634img_df[1:5, 1:5]# A tibble: 5 × 5
x `1` `2` `3` `4`
<int> <dbl> <dbl> <dbl> <dbl>
1 1 0.914 0.914 0.914 0.910
2 2 0.929 0.929 0.925 0.918
3 3 0.910 0.910 0.902 0.894
4 4 0.906 0.902 0.898 0.894
5 5 0.898 0.894 0.890 0.886Don’t be Cruel
tmp <- img_df |> select(-x)
dim(tmp)[1] 800 633tmp[1:5, 1:5]# A tibble: 5 × 5
`1` `2` `3` `4` `5`
<dbl> <dbl> <dbl> <dbl> <dbl>
1 0.914 0.914 0.914 0.910 0.902
2 0.929 0.929 0.925 0.918 0.910
3 0.910 0.910 0.902 0.894 0.886
4 0.906 0.902 0.898 0.894 0.890
5 0.898 0.894 0.890 0.886 0.882# Scaled and centered
tmp_norm <- scale(tmp, center = TRUE, scale = TRUE)
tmp_norm[1:5, 1:5] 1 2 3 4 5
[1,] 0.3062542 0.3271632 0.3352950 0.3137665 0.2662779
[2,] 0.4172912 0.4326572 0.4124186 0.3645892 0.3165605
[3,] 0.2784949 0.3007897 0.2581714 0.2121212 0.1657127
[4,] 0.2507356 0.2480427 0.2324635 0.2121212 0.1908540
[5,] 0.1952171 0.1952957 0.1810477 0.1612985 0.1405714# Covariance matrix
cov_mat <- cov(tmp_norm)
dim(cov_mat)[1] 633 633cov_mat[1:5, 1:5] 1 2 3 4 5
1 1.0000000 0.9945150 0.9877773 0.9848553 0.9820028
2 0.9945150 1.0000000 0.9966495 0.9902557 0.9834383
3 0.9877773 0.9966495 1.0000000 0.9958584 0.9866195
4 0.9848553 0.9902557 0.9958584 1.0000000 0.9931752
5 0.9820028 0.9834383 0.9866195 0.9931752 1.0000000Wooden Heart
Doing the PCA manually
# Decomposition/Factorization into eigenvalues and eigenvectors
cov_eig <- eigen(cov_mat)
names(cov_eig)[1] "values" "vectors"# Eigenvalues (variances)
cov_evals <- cov_eig$values
cov_evals[1:5][1] 231.41622 120.60984 56.89806 31.05158 22.82532# Eigenvectors (principal components)
cov_evecs <- cov_eig$vectors
cov_evecs[1:5, 1:5] [,1] [,2] [,3] [,4] [,5]
[1,] -0.006164854 -0.06573463 0.02875530 -0.03933564 0.06010992
[2,] -0.006734353 -0.06614096 0.02858038 -0.03849782 0.05901673
[3,] -0.007154980 -0.06590388 0.02736392 -0.03892858 0.05852467
[4,] -0.007471463 -0.06604222 0.02671015 -0.03827302 0.05944892
[5,] -0.006484754 -0.06614490 0.02788219 -0.03994717 0.06062967# Rotation matrix -- ie the coordinates of the
# original data points translated into the
# transformed coordinate space prcomp$rotation
tmp_rot <- tmp_norm %*% cov_evecs
dim(tmp_rot)[1] 800 633tmp_rot[1:5, 1:5] [,1] [,2] [,3] [,4] [,5]
[1,] 11.589359 -4.479811 -2.268584 3.666006 4.655793
[2,] 10.917918 -4.386686 -3.477785 4.237974 6.417180
[3,] 9.411595 -4.239953 -4.511044 3.983355 6.585201
[4,] 8.768736 -3.946033 -4.653053 3.398185 4.503424
[5,] 8.452248 -3.153785 -4.491075 2.568274 1.609303# Should be zero
round(cov(cov_evecs), 2)[1:5,1:5] [,1] [,2] [,3] [,4] [,5]
[1,] 0 0 0 0 0
[2,] 0 0 0 0 0
[3,] 0 0 0 0 0
[4,] 0 0 0 0 0
[5,] 0 0 0 0 0Clean up your own Backyard
img_pca <- img_df |>
select(-x) |>
prcomp(scale = TRUE, center = TRUE)
pca_tidy <- tidy(img_pca, matrix = "pcs")
pca_tidy# A tibble: 633 × 4
PC std.dev percent cumulative
<dbl> <dbl> <dbl> <dbl>
1 1 15.2 0.366 0.366
2 2 11.0 0.191 0.556
3 3 7.54 0.0899 0.646
4 4 5.57 0.0491 0.695
5 5 4.78 0.0361 0.731
6 6 4.56 0.0328 0.764
7 7 4.06 0.0261 0.790
8 8 3.66 0.0212 0.811
9 9 3.37 0.0179 0.829
10 10 3.28 0.0170 0.846
# ℹ 623 more rowsAll Shook Up
names(img_pca)[1] "sdev" "rotation" "center" "scale" "x" What are these? sdev contains the standard deviations of the principal components. rotation is a matrix where the rows correspond to the columns of the original data, and the columns are the principal components. x is a matrix containing the value of the rotated data multiplied by the rotation matrix. Finally, center and scale are vectors showing the centering and scaling for each observation.
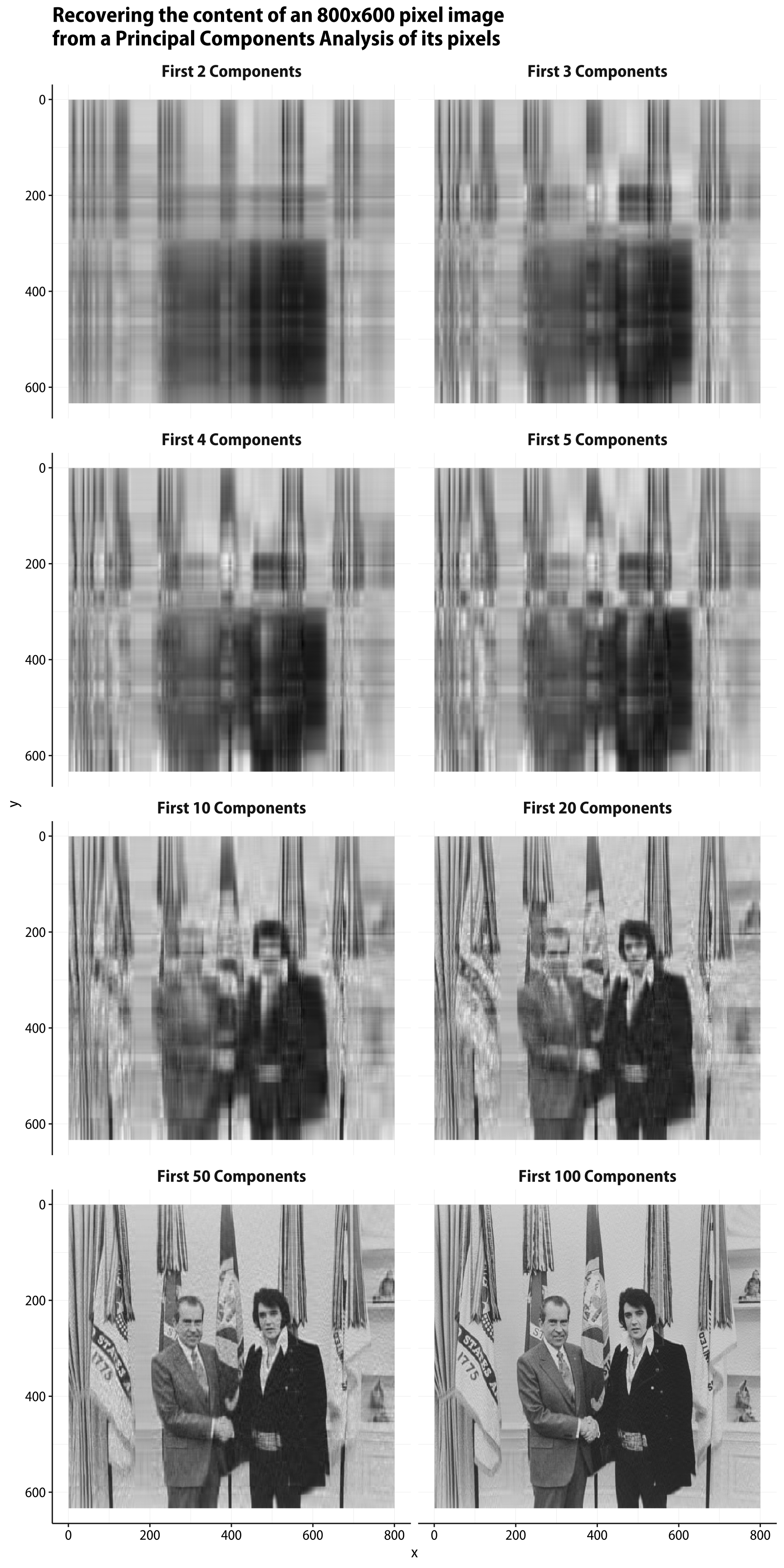
Now, to get from this information back to the original data matrix, we need to multiply x by the transpose of the rotation matrix, and then revert the centering and scaling steps. If we multiply by the transpose of the full rotation matrix (and then un-center and un-scale), we’ll recover the original data matrix exactly. But we can also choose to use just the first few principal components, instead. There are 633 components in all (corresponding to the number of rows in the original data matrix), but the scree plot suggests that most of the data is “explained” by a much smaller number of components than that.
I Gotta Know
Here’s a function that takes a PCA object created by prcomp() and returns an approximation of the original data, calculated by some number (n_comp) of principal components. It returns its results in long format, in a way that mirrors what the Imager library wants. This will make plotting easier in a minute.
reverse_pca <- function(n_comp = 20, pca_object = img_pca){
## The pca_object is an object created by base R's prcomp() function.
## Multiply the matrix of rotated data by the transpose of the matrix
## of eigenvalues (i.e. the component loadings) to get back to a
## matrix of original data values
recon <- pca_object$x[, 1:n_comp] %*% t(pca_object$rotation[, 1:n_comp])
## Reverse any scaling and centering that was done by prcomp()
if(all(pca_object$scale != FALSE)){
## Rescale by the reciprocal of the scaling factor, i.e. back to
## original range.
recon <- scale(recon, center = FALSE, scale = 1/pca_object$scale)
}
if(all(pca_object$center != FALSE)){
## Remove any mean centering by adding the subtracted mean back in
recon <- scale(recon, scale = FALSE, center = -1 * pca_object$center)
}
## Make it a data frame that we can easily pivot to long format
## (because that's the format that the excellent imager library wants
## when drawing image plots with ggplot)
recon_df <- data.frame(cbind(1:nrow(recon), recon))
colnames(recon_df) <- c("x", 1:(ncol(recon_df)-1))
## Return the data to long form
recon_df_long <- recon_df |>
tidyr::pivot_longer(cols = -x,
names_to = "y",
values_to = "value") |>
mutate(y = as.numeric(y)) |>
arrange(y) |>
as.data.frame()
tibble::as_tibble(recon_df_long)
}It’s Now or Never
## The sequence of PCA components we want
n_pcs <- c(2:5, 10, 20, 50, 100)
names(n_pcs) <- paste("First", n_pcs, "Components", sep = "_")
## Map reverse_pca()
recovered_imgs <- map(n_pcs, reverse_pca) |>
list_rbind(names_to = "pcs") |>
mutate(pcs = str_replace_all(pcs, "_", " "),
pcs = factor(pcs, levels = unique(pcs), ordered = TRUE))
recovered_imgs# A tibble: 4,051,200 × 4
pcs x y value
<ord> <dbl> <dbl> <dbl>
1 First 2 Components 1 1 0.902
2 First 2 Components 2 1 0.902
3 First 2 Components 3 1 0.902
4 First 2 Components 4 1 0.899
5 First 2 Components 5 1 0.892
6 First 2 Components 6 1 0.886
7 First 2 Components 7 1 0.877
8 First 2 Components 8 1 0.858
9 First 2 Components 9 1 0.823
10 First 2 Components 10 1 0.772
# ℹ 4,051,190 more rowsJailhouse Rock
ggplot(data = recovered_imgs,
mapping = aes(x = x, y = y, fill = value)) +
geom_raster() +
scale_y_reverse() +
scale_fill_gradient(low = "black", high = "white") +
facet_wrap(~ pcs, ncol = 2) +
guides(fill = "none") +
labs(title = "Recovering the content of an 800x600 pixel image\nfrom a Principal Components Analysis of its pixels") +
theme(strip.text = element_text(face = "bold", size = rel(1.2)),
plot.title = element_text(size = rel(1.5)))